Abstract
All aspects of life activities in living cells are mediated/executed and regulated by a vast number of networks, comprising a wide spectrum of components, starting with simple biomolecules and ending with the whole organism, and functioning within a precisely organized tight framework. Proper mediation of cellular activities necessitates their inclusion within the context of structured and organized network systems capable of regulating/coordinating and synchronizing the countless numbers of biological processes occurring within living cells. The number of biological networks and pathways within the living cell is considerably huge, being dependent on the structural complexity and functional capabilities of the cell. Pathogenesis and progression of human diseases result from functional disturbances of biological networks within the cell as disturbed network function leads to deleterious effects on physiological processes dependent on, and mediated by, affected network(s). Ensuing pathological processes, defined by the nature of disturbed networks and the specific organs or tissues affected, pave the way for the development of pathognomonic and characteristic disease entities. As most network functions are dependent on relatively small number of key regulatory biomolecules, i.e. enzymes/proteins and signal transducing factors, it follows that functional disturbances of biological networks and pathogenesis of disease states can be attributed, in most instances, to quantitative and/or qualitative abnormalities of these key regulatory molecules. Study and analysis of the structural designs and the functional mechanisms of biological networks would have crucial and important impacts on many theoretical and applied aspects of biology, in general, and of medical sciences in particular. Meticulous study of biological networks represents an important and integral aspect in study of biology. Interpretation and analysis of key information deduced from observing and analyzing structural designs and functional characteristics and dynamics of biological networks discloses and defines the basic framework within which life activities in living cells are initiated, adapted to physiological requirements, maintained, and terminated upon completion of their aims. More important, however, is the contribution of this information to proper understanding of the different mechanisms responsible for regulating and synchronizing the functions and performances of the vast spectrum of different network categories within the cell. In addition to its vital scientific significance, discovering and defining the key pivotal structural and regulatory molecules within life-mediating networks, and along different pathways responsible for controlling functional dynamics of the network, represent an indispensable diagnostic approach insistent for designing proper therapeutic approaches to diseases caused by network defects.
Author Contributions
Academic Editor: Bobbie-Jo M, Webb-Robertson, Senior Research Scientist, Pacific Northwest National Laboratory, Computational Biology and Bioinformatics, Richland, WA , USA.
Checked for plagiarism: Yes
Review by: Single-blind
Copyright © 2018 Mohammad Saad Zaghloul Salem
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
The authors have declared that no competing interests exist.
Citation:
Contents
1. Introduction
2. Types of network systems
A. Natural networks
B. Artificial networks
C. Biological networks
3. Biological networks
4. Characteristic features of biological networks
5. Design of biological networks
6. Structural and functional distortion of biological networks
7. Mechanisms of regulation of biological networks
7.1. Regulatory roles of ribozymes and riboswitches
7.2. Regulation of higher genomic regulatory networks
8. Types and classification of biological networks
9. Temporal classification of biological networks
10. Functional classification of biological networks
10.1. Genome preserving networks
10.2. Transcription regulatory networks or transcriptome networks
10.3. Translation regulatory networks or proteome networks
10.4. Developmental networks
10.4.1. Characteristics and regulation of developmental networks
10.4.2. Developmental signaling pathways
10.5. Signal transduction networks
10.5.1. Characteristics of signal transduction mechanisms
10.5.2. Important signal transduction networks
10.6. Apoptosis networks
10.6.1. Cascade of signaling mechanisms of apoptosis
10.6.2. Characteristic features of apoptosis
10.7. Oscillatory rhythm networks
10.7.1. Circadian rhythm networks
10.7.2. Intrinsic rhythm networks
10.8. Executive networks
10.9. Metabolic networks
10.9.1. Functional regulation of metabolic networks
10.9.1 A. Quantitative regulation of functions of metabolic networks
10.9.1 B. Qualitative regulation of functions of metabolic networks
10.9.2. Metabolic disorders
10.9.2 A. Congenital metabolic disorders
10.9.2. B. Acquired metabolic disorders
10.9.3. Pathogenesis of metabolic disorders
10.9.4. Classification of metabolic disorders
10.10. Preserving and repair networks of the genome, the transcriptome and the proteome
10.10.1. Anti-mutation networks of the genome
10.10.2. Transcriptome preserving and repair networks
10.10.3. Proteome preserving and repair networks
10.11. Neuronal brain networks
11. Proposed hypothesis of mechanisms of function of neuronal networks
12. Challenges to defining key regulatory molecules of biological networks
13. Importance of studying and analysis of biological networks
14. References
1. Introduction
A network is an organized deterministic system composed of many structural components, units and sub-units, working in a predefined programmed and recurring manner to perform specific function(s) according to regulatory rules controlling functions/responses/interactions and cooperation of its components. There are many different kinds of network systems, for example engineered networks, electrical networks, mechanical networks, industrial networks, communication networks, natural ecological networks and biological networks. Each kind of network systems is constructed and controlled according to system-specific standards to perform the required function(s). Biological networks are organized deterministic systems that constitute the framework of life activities of the whole structural spectrum of living matter that includes biomolecules, e.g., nucleic acids and proteins, cell organelles, e.g., mitochondria and cytoskeleton, whole cells, tissues, organs and, at last, the whole organism (Figure 1 & Figure 2).
Figure 1.The design of biological networks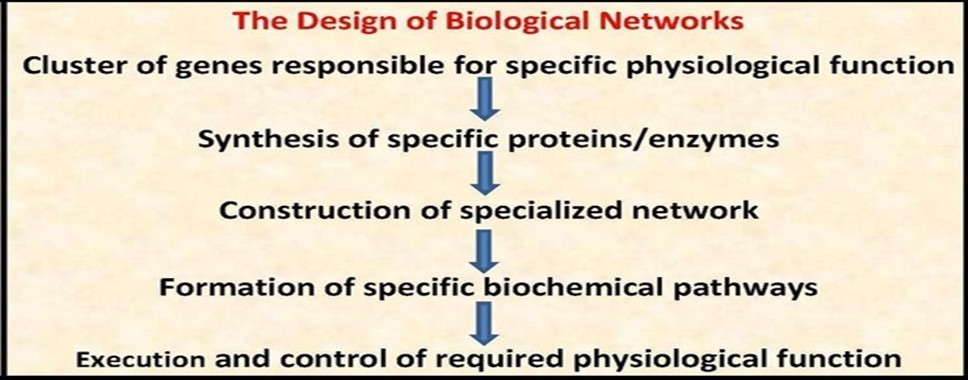
Figure 2.The concept of biological/biochemical pathways
The concept of the network is a universal basic frame-work within which the actions/ reactions/interactions of related objects or components of existing systems exert their activities. Moreover, it also regulates interactions between different groups of apparently un-related systems. For example, apart from being a component of the cosmic networks that regulate the behavior of similar systems in the milky way galaxy, the specific networks of the solar system responsible for regulating too many parameters concerning heat production and light emission by the sun are intimately related to a large number of vital networks responsible for regulating biological activities of nearly all living creatures on the earth. Although the rules that govern these interactions between apparently unrelated networks are not known, the effects of the networks responsible for initiating/maintaining/regulating heat production in the sun have a crucial impact on all network systems on the earth including biological networks regulating the life of human beings, animals, plants, and other simpler organisms as well as physical networks that regulate light and temperature input and distribution allover all solid parts and water stores of the earth.
In spite of the marked differences between different types of networks, they share common features including a basic model design reflecting composition and assemblage of variable numbers of structural units, each consisting of a number of nodes organized into modules. The modular design of the network characterizes specialized functional units coordinated to regulate and control the behavior of the network. Other shared features between different network systems include presence of key regulatory components, continuity of network action(s) as long as balance between input data/substrates and output data/products is maintained, preservation of network performance provided that internal/external optimal functioning conditions are conserved, and presence of stringent surveillance and control modules to optimize network function(s) and, in some instances, to minimize possible hazards that may result from malfunctioning of any unit or component of the network.
A comprehensive architectural design of key structural units and key regulatory components of any/all artificially structured/engineered networks is a prerequisite and a predefined characteristic of the network system. However, the same does not apply to either natural networks or biological networks. In natural networks that constitute the framework of actions and interactions of natural ecological systems, many key structural units and key regulatory components of these networks are, largely, speculative. This is attributed to the fact that a considerable proportion of the spectrum of natural forces that constitute and control natural phenomena is, still, ill defined. Biological networks, probably, are the least as regards availability of sufficient and satisfactory interpretation of data concerning their structural designs, their functional capabilities, their regulatory units, and the mechanisms of synchronization and coordination of their components. In spite of the continuing gush of tremendous sources of information in the field of biology, too little is known about the exact nature, the functional potential, the hierarchical organization and the flexibility of responses to external effectors of the extremely wide spectrum of network systems encompassing and controlling the innumerable groups of biological networks responsible for mediating the countless numbers of cellular life activities in both physiological as well as pathological conditions of human and other living organisms.
However, it must be emphasized that a considerable difficulty in study of biological networks from a medical point of view resides in the fact that analysis of network systems in general is carried out by theorists in physics and mathematics who use advanced and complex mathematical analysis and algorithmic approaches for constructing computational design models of network systems that fit, primarily, to artificially engineered networks. Additionally, technical terminology used within this context does not coincide with the more simple descriptive terminology used for characterization of biological processes. For example, the definition of the network as a (set of nodes and a set of directed or undirected edges between the nodes) seems quite ambiguous, or even meaningless, from a medical/genetic/physiological point of view that describes biological networks in a more simpler, descriptive and applicable terms, e.g., a biological network is an organized system composed of a number of proteins/enzymes/other molecules that act in a coordinated pattern within a specific cellular compartment to perform specific physiological functions.
Difficulties in discovery and characterization of biological networks, whether individually or as clusters of functionally related networks, are related to the colossal wide spectrum of the variable structural components of these networks that comprises thousands of genes, probably similar numbers of different categories of RNAs, and much more larger numbers of different types of proteins. Advances in molecular diagnostic techniques represented a major breakthrough in trials to disclose and define a large portion of these components, and diagnostic approaches based on more advanced analytical methods, e.g., quantum dynamics and nano-dynamics based techniques and computational genomics, are expected to offer greater diagnostic capabilities in this regard.
2. Types of Network Systems
Classification of network systems is not an easy task because of the multiple and different criteria that define and characterize each type of network systems. Examples of such criteria include, for instance: structural components, motif patterns, hierarchical design, modularity, robustness, perception and sensitivity to external stresses, redundancy of components, functional performance, automation, scale-free nature of the network, in addition to many other criteria. Accordingly, many different categories of networks could be identified and delineated depending on the particular criteria used for classification. However, it must be emphasized that no satisfactory or a globally accepted classification scheme of networks is agreed upon, and that the classification framework of networks presented here is a tentative suggested approach taking into consideration the basic features that characterize each type. Within this context, three distinct categories of networks, with many subcategories of each, could be identified and delineated:
A. Natural Networks or naturally existing ecological systems that constitute, define and regulate the framework of all and varying aspects of earth ecology like climate networks, animal-plant and microbial biome networks and aquatic networks.
B. Artificial Networks that are designed, constructed, engineered and structured for specific purposes like electrical networks, industrial networks and computer-informatics networks.
C. Biological Networks that mediate and regulate all life activities of living organisms. Examples of biological networks include genomic networks, proteome networks, metabolic networks, regulatory networks, neuronal networks, inter-cellular networks and many others.
3. Biological Networks
The basic essence of the concept of biological networks resides in the fact that all aspects of life activities in living cells are mediated/executed and regulated by a vast number of networks, comprising a wide spectrum of components, starting with simple biomolecules and ending with the whole organism, and functioning within a precisely organized tight framework. On the cellular level, all life activities are precisely and tightly connected and interconnected in many different specific programmed predetermined mechanisms, e.g., cascade-loop reactions/stimulus-response reactions/threshold-effect reactions and feedback-loop reactions. Proper mediation of cellular activities necessitates their inclusion within the context of structured and organized network systems capable of regulating/coordinating and synchronizing the countless numbers of biological processes occurring within living cells (Figure 3).
Figure 3.Genetic regulation of life activities and the concept of biological networks
The number of biological networks and pathways within the living cell is considerably huge, being dependent on the structural complexity and functional capabilities of the cell. Each network/pathway performs a specific physiological function within the global frame of cellular activities. The biological frame of life activities encompasses three major aspects in living organisms:
1.Keeping the vital structural design and integrity of different cellular components and cell organelles, e.g., cell nucleus and sub-nuclear structures, cell cytoskeleton, mitochondria, cell membranes, Golgi body, endoplasmic reticulum, cell pores and transport channels and intercellular connections etc.
2.Maintaining optimal functional performance of the whole spectrum of cellular activities including cell growth, division, proliferation, differentiation, migration, apoptosis and single and multicellular responses to, and interactions with, environmental effectors.
3.Integrating andcomplementing functions of all components of multicellular and higher organisms, cells-tissues-organs, to ensure optimal biological fitness of the organism.
Pathogenesis and progression of human diseases result from functional disturbances of biological networks within the cell. Disturbed network function leads to deleterious effects on physiological processes dependent on, and mediated by, affected network(s). Ensuing pathological processes, defined by the nature of disturbed networks and the specific organs or tissues affected, pave the way for the development of pathognomonic and characteristic disease entities. As most network functions are dependent on relatively small number of key regulatory biomolecules, i.e. enzymes/proteins and signal transducing factors, it follows that functional disturbances of biological networks and pathogenesis of disease states can be attributed, in most instances, to quantitative and/or qualitative abnormalities of these key regulatory molecules.
4. Characteristic Features of Biological Networks
In spite of the remarkable differences between individual network categories, and even between secondary networks and sub-pathways within each category as regards its structural design and its functional spectrum, biological networks, in general, have and share common characteristic dynamic features determined by natural laws of thermodynamics. These features include, for example, basic similar dynamic mechanisms responsible for initiation of reaction cascades of different components of the network and synchronization of these cascades for multistage networks, maintaining rate limiting reactions at optimal performance levels, synchronization of network function(s) with other functionally related networks or pathways, ensuring efficient and optimal balance between substrate input and product output of the network, maintaining optimal and efficient loop of supply-flow-consumption of energy by the network, adapting network performance to physiological needs and to emergent changes in reaction environment and, finally, termination of reactions and interactions of different pathways of the network after attaining its predefined targets and performing its functions.
Study, analysis, and characterization of biological networks must take into consideration a major difference that characterizes this category of networks from other types of network systems; viz. biological networks are living systems and not merely constructed materialistic systems designed for performing specific functions. Many significant consequences of the living nature of biological networks follow this delineation, and can be summarized in the following observations:
4.1. Though biological networks are constructed from living units and subunits composed of biomolecules that show limited qualitative variations being composed of structural proteins, catalytic enzymes, ribozymes, and nucleic acids, they show wide quantitative variations from small local intracellular networks composed of few biomolecules, e.g., metabolic networks mediating enzymatic conversion of one compound to another, to exceedingly large networks composed of large numbers of biomolecules exerting their functions across many tissues and organs, or even on the level of the whole organism, like hormonal and neuronal networks.
4.2. The unique nature of the living design of biological network systems imparts to them peculiar characteristics that differentiate and characterize their performance from that of artificially structured network systems, irrespective of the apparently wider quantitative structural complexity of artificial networks. The differences stem from differences in flexibility to external stresses due to differences in modularity design of the two network systems, existence of alternative pathways for possible functional defect in any of the units/subunits composing the system, and presence of unique dynamic regulatory mechanisms exerted and controlled by two major, among many other, life preserving systems; viz. the neural and the endocrinal systems.
4.3. A crucial feature of biological networks resides in their ability to anticipate, and interact to, possible causes of performance inefficiency of the network due to many reasons like changes in input, e.g., nutritional deficiency, or changes in surrounding environment, e.g., infection. These predictive and corrective responses are initiated and conducted by many different mechanisms. Sensing networks that act in precise synchrony and cooperation with functionally related executive networks, constitute major life preserving measures of utmost importance for living organisms. For instance, sensory neuronal and metabolic networks responsible for sensing hypoglycemia, hypocalcemia or hypothermia, and the functionally related executive networks responsible for, and responding by, enhancing glycogenolysis, increasing blood calcium level and ATP production, respectively, reveal this crucial feature which reflects the wide range of flexibility of biological networks to changing external and/or internal effectors and working conditions.
4.4. The round the clock continuous dynamic changes of the input/output parameters and conditions of life activities of living organisms necessitates presence of precise master systems responsible for surveillance, detection and correction of any ensuing defects in structural design or functional performance of the network. Examples of these master regulatory networks include DNA/RNA/Protein proofreading and repair network systems, e.g., DNA repair enzymes, guide RNA and chaperones. These real time dynamic responses of biological networks to existing and to evolving changes are indispensable for maintaining vital homeostasis needed for proper regulation of life processes in living cells, as well as for prevention of occurrence or progression of detrimental errors that can lead to pathogenesis of disease.
4.5. The pivotal role played by enzymes in mediation and regulation of all biological networks in living organisms, without exception, has no counterpart in artificially constructed networks. Though many types of natural networks that shape and control main ecological bio-systems of the earth involve actions of enzymes, many natural networks do not involve enzymes as part of their design, and those that do involve it as an implicit component of some of its constituent units.
The crucial roles attributed to enzymes in constructing, mediating and regulating functional performance of biological networks resides in their peculiar ability to compose/decompose/change/reshape/locate/relocate other simple and complex biomolecules in a unidirectional as well as in a bidirectional manner within reaction environment as well as within cellular components hosting these reactions. This feature of enzyme action is responsible for the exquisite precision and efficiency of functional performance of biological networks. In addition, an important feature of enzyme dynamics that have crucial roles in network functions is the ability of enzymes to undergo transient association into multifaceted, more stable functional complexes that lead to marked increase in functional efficiency of the network. The presence of multiple functionally related isoforms of most enzymes involved in metabolic regulation represents a protective mechanism to preserve the integrity of the metabolic network in the face of mutations that can cause defective or deficient synthesis of one of the isoforms. Another major feature of enzyme dynamics allowing them to act as central regulatory/executive units of biological networks is their rapid, sometimes, instantaneous responses to changes in performance requirements. Enzymes act as catalysts that allow biochemical reactions to proceed, and allow rapid regulation of metabolic pathways in response to changes in the cell's environment or extrinsic signaling input from other cells, because enzymes exist as preexisting components, the activation of which is a much faster process compared to activation/regulation of gene expression for synthesis of required enzyme proteins.
4.6. An important feature of biological networks that might have important implications in diagnosis and treatment of many diseases is their conformation, as a complex organized system, to some rules different from, sometimes even contradictory to, some basic rules of artificial network dynamics. This argument debates the alleged assumption that ordered systems are generally less controllable/regulated than disordered ones. For instance, the hypothesis that this assumption could be applied to cancer therapy trials 1 is obviously unreasonable, because progression of carcinogenesis reflects ongoing decomposition of the cellular systems of malignant cells, i.e. progression from order to more disorder and basic clinical facts of cancer therapy clearly shows that treatment of early cancer, i.e. relatively more ordered system, is much more effective than later intervention when cancer cell systems become more disordered, due to involvement of more cellular components and networks in development of the malignant phenotype, thus adding more disorder to the system.
4.7. Most cellular networks are confined to the intracellular environment, even to specific cell organelles either individually or in concert to accomplish specific physiological processes, e.g., metabolic networks responsible for fatty acid oxidation within the mitochondria and regulatory/proofreading networks responsible for post-translation modifications of proteins during their sequential passage along the endoplasmic reticulum and Golgi apparatus. However, biological networks might comprise distant units and subunits dispersed within many tissues and organs, that are tightly interconnected, synchronized and functionally coordinated via neural/endocrinal regulatory motifs. For example, preservation of blood and body fluid osmolality has a critical impact on life activities of cells and its precise regulation is attained through a crucial water homeostasis network comprising the brain, the kidney, the skin, the heart and the eyes as well. In pathophysiological conditions causing hyper-osmolality states, physiological alterations due to sensation of thirst/diminished water content of skin/decreased intraocular pressure due to diminished secretion of intraocular fluid/diminished volume sensation of auricles and ventricles of the heart/diminished pressure in blood vessels due to fall in blood pressure, all trigger local signaling pathways in affected dehydrated cells and tissues. Network modules responsible for maintaining network function, i.e. preservation of proper blood/fluid osmolality, begin to exert their corrective roles via different processes, e.g., stimulation of osmo-receptor nodes of neural networks of the hypothalamus responsible for synthesizing the prepro-form of vasopressin, or antidiuretic hormone, and for controlling its conversion to the mature form and its release by the posterior pituitary. Other hormones taking part in maintaining water homeostasis and participating in regulating water excretion by the kidney and prophylaxis against pathological consequences of plasma hyper-osmolality, e.g., renin/aldosterone/secretin/aquaporins, might also constitute additional/alternative/subsidiary subunits or modules of the water homeostasis network, and are affected in a similar way. Following these responses, vasopressin and other hormones begin to exert their corrective functions to restore network performance and function(s) according to their predefined roles, e.g., affecting permeability of renal tubules to water, augmenting sensitivity of collecting ducts to vasopressin and the like.
4.8. A crucial feature that characterizes biological networks is the marked degree of flexibility of performance of the network in response to changes of functioning conditions, and to a lesser extent, to changes involving structural units, e.g., biomolecules and organelles of the network. For instance, flexibility of performance of artificially engineered networks is limited by the predefined design features of the network, and unexpected external stress or change of functioning conditions could be tolerated up to a limit followed by failure of network function. Though the traditional definition of chaos or deterministic chaos states that in absence of random events, the future behavior of deterministic systems is determined by their initial conditions, predictability of this behavior is not possible or, at least, hard to measure. The question of whether biological systems, including the genome, the proteome, the transcriptome and other biome networks, can be dealt with as deterministic systems or not has no definite or universally accepted answer. On one hand, in contrast to the modularity of engineered networks that have no or limited performance flexibility in response to stress, biological networks are fully flexible networks. Even with unexpected random events, e.g., external environmental mutagenic effects, these systems have a precise degree of predictability of their behavior, e.g., DNA repair mechanisms. Many factors underlie functional flexibility of biological networks. First, presence of multiple regulatory modules, nearly of all parts and along all reaction steps of the network to detect early malfunctioning signs of the network. Second, existence of varying and different corrective processes triggered by signaling pathways and conducted by efficient functional responses of major components of the network system, e.g., the genome, the proteome and the transcriptome.
4.9. An important feature of many biological networks is the multifunctionalcapabilities ofsome of its key components. For instance, many proteins and enzymes that constitute parts of important regulatory/signaling networks have many functions and can regulate or affect many other components/pathways of the network. A prominent example is Glyceraldehyde 3-phosphate dehydrogenase, (GAPDH or G3PDH), an enzyme involved in glycolysis and energy production through break down of glucose. In addition to this metabolic function, GAPDH has been found implicated in performing other non-related physiological functions including neuronal transmission, intracellular trafficking of translated proteins, transcription activation, iron homeostasis and initiation of apoptosis.
Though the traditional interpretation of this multifunctional property of enzymes/proteins rests mostly on the presence of multiple functional domains each capable of mediating a specific function, other plausible interpretations could, also, be assumed, e.g., binding to another protein/enzyme thus forming a novel functional catalytic component, structural conformational change thus offering more functional options like binding to membranes/cell pores/cytoskeleton regions, or even degradation or disassembly to smaller different but, still, functionally specialized units.
A relatively recent postulation attributes this multifunctional property to a vague hypothesis termed gene sharing, i.e. a single gene is sharing in performing multiple/different/unrelated functions via its encoded protein product 2. The prominent examples of proteins within this context are the moonlighting proteins. Though the main function of moonlighting proteins is enzymatic catalysis, they also serve other non-enzymatic functions in view of their many functional categories which include membrane receptors, ion channel regulators, chaperones and initiators of apoptosis 3.
Meticulous analysis of the gene sharing hypothesis, however, reveals that the essence of the hypothesis seems unreasonable because it is based on unnecessary theoretical postulations trying to deduce new concepts to explain findings already interpretable by the traditional dogmas of biochemistry and physiology. The postulation argues that most, if not all, proteins perform a variety of functions in the same and in different species, and that this is a fundamental necessity for evolution. However, the cardinal examples of the hypothesis, crystallins and actin, that have diverse unrelated functions are not unique in this respect, as multitudes of proteins already have this multifunctional property and perform diverse unrelated functions including regulation of gene expression, e.g., p53 4, fragile X mental retardation protein (FXMR1) 5, and many others.
The Gene sharing hypothesis represents another desperate attempt to support the irrational concept of evolution by postulating unreasonable assumptions that have no logical support. More advanced analytical measures will, probably, reveal that the underlying molecular mechanisms of this multifunctional property of proteins/enzymes are mediated, and can be interpreted within the context of the aforementioned classic dogmas of biochemistry and physiology.
4.10. A hierarchical master regulatory system responsible for controlling network function(s) probably exists for most, if not all, biological networks within living cells. Though the exact mechanisms through which this presumed control system exerts its effects are vaguely defined, a tentative hypothetical frame based on accumulating observations might be postulated in this regard. Within the context of this postulated hypothesis, all biological networks in the cell are considered as subordinate entities constructed and organized according to the information embodied within the genome. The genome, as the prime entity in this hierarchical system, defines all types of networks needed for mediating all life processes in the cell. Genome function(s), in turn, are probably regulated by genomic regulatory networks composed of, and controlled by, different constituents, e.g., microRNA/histones/proteins, that regulate and maintain structural and functional integrity of the genome. These genomic regulatory networks mediate critical tasks encompassing a wide variety of genome functions and behavior like regulation of DNA replication/regulation of DNA repair/regulation of cell division, and many others.
4.11. In contrast to the modularity of artificially engineered networks, that have no or limited performance flexibility in response to external stresses or to ensuing changes in internal performance, biological networks are fully flexible networks. Even with unexpected random events, e.g. external mutagenic effects, these systems have a precise degree of predictability of their behavior, e.g. DNA repair mechanisms. Sometimes, recognition or detection of this flexibility in behavior is blurred upon looking to the effects of mutation in general. Mutation-induced genetic disorders should not, and could not, be considered as chaotic behavior, in the scientific sense, of the genome. Rather, they reflect and represent precisely predefined behavior in response to random events determined by the initial conditions of the system.
4.12. Although the genome defines the proteome that constitutes the structural components of the majority of developmental/metabolic/ signaling networks all through the life cycle of the cell, auto-regulatory subsidiary circuits and modulator pathways, composed of protein by-products of a network, can mediate some integral functions within the mother network. Self-exerted auto-regulation, mediated by a wide variety of biomolecules including proteins/enzymes/microRNAs, is a critical mechanism that seems to plays an important role in regulating and fine tuning the function of most biological networks. These functions include, for instance, rate-limiting effects/feedback potentiation or inhibition/signaling and synchronization, among other functions. The source as well as the behavior of this postulated inherent auto-regulation exerted by proteome components on biological networks, including even genomic regulatory networks, has no clear interpretation. Postulations that attribute this behavior to mere conformation with biochemical/biophysical laws ruling the network/reaction environment can, partly, offer an explanation but they can not interpret the persistence of this behavior in different pathophysiological conditions. Regulatory protein by-products of parent networks are, obviously, out of control of the genome as they are not synthesized by genes, and their apparent roles might be controlled by information embodied within the proteome, rather than the genome, of the cell.
4.13. The genome of the cell defines and controls synthesis of its proteome through a strict precise hierarchical pathway system including many networks that act in accurate temporal sequence to regulate gene function via gene activation and start of transcription, post-transcription modifications of mRNA, translation, post-translation modifications of synthesized proteins and, finally, intracellular/extracellular trafficking and localization of synthesized proteins. Following the final structural organization of the proteome of the cell, intracellular networks regulating vital processes in the cell are formed. These networks comprise the vast majority of executive cellular networks responsible for energy production/maintenance of optimal physical characteristics of the cytosol/preservation of the proper balanced biochemical composition of all cellular components/regulation of metabolism/regulation of transport across cell membrane and membranes of cell organelles/synthesis, construction and maintenance of cytoskeleton components of the cell/regulation of excretory processes/regulation of secretory processes/regulation of apoptosis and regulation of all other vital processes required to maintain proper and optimal performance of life activities and cellular functions.
The last level of the hierarchical organization of cellular networks within this context of gene function includes networks responsible for regulating intercellular and extracellular processes. Intercellular networks regulate contact and communication between cells, synchronization of cellular responses to changes in extracellular microenvironment and global mass cellular behavior like cell growth, cell differentiation, cell migration and apoptosis. Similarly, extracellular regulatory networks control vital processes necessary for homeostasis of life environment of multicellular organisms including global mass cellular behavior like cell growth, cell differentiation, cell migration and apoptosis during embryonic and fetal development, regulation of cardiac output, blood pressure and local perfusion of tissues, regulation of blood osmolality and electrolyte balance, regulation of neuronal transmission, regulation of immune defense mechanisms like quorum sensing and mass cellular aggregation in inflammatory conditions, regeneration and differentiation of stem cells in certain pathological states, and regulation of an endless list of other vital functions in multicellular organisms.
4.14. Study and analysis of biological networks in accordance to the classic definition of network systems has to recall and put into consideration additional peculiar features that characterize some subcategories of these networks. Some types of biological networks, mostly metabolic pathways, are temporarily formulated and constructed to perform specific functions, e.g., metabolism of drugs or other administered foreign substances. Following performing the required task(s), the network is disassembled to its component units/subunits. This fate is important to save energy, recycle network components for other purposes and to avoid possible conflict with other pathways. Accordingly, large numbers of biological networks have no enduring or permanent structural framework, they ate formulated and constructed to face temporary or contingent specific cellular requirements. The significance of this feature raises many questions regarding the functional potential of the genome/proteome profile of the cell. For instance: are there hidden networks and pathways that appear only upon need, e.g., upon exposure to extrinsic stressful effects, and if so, are there predefined/preprogrammed genomic/genetic registry data ready for transcription/translation upon need. This feature denotes the unlimited reactive potential and the countless synthetic capabilities of the genetic material in response to environmental stresses and offers an explanation to the existence and persistence of huge numbers of living organisms in spite of unfavorable environmental conditions. The temporary nature of this phenomenon contradicts its consideration within any evolutionary context as it does not become a consistent fixed part of the life frame of the organism.
4.15. Although most aspects of the postulated hierarchical system responsible for regulating functions and interactions of biological networks are well defined on cellular level, little is known as regards its mechanisms of action on the whole organism level. In humans and other multicellular organisms, two major regulatory systems, viz. the nervous and the endocrinal systems, exert coordinated and synchronized regulatory functions though via different mechanisms. A master regulatory hierarchical system dominating both these systems does not exist, and hypotheses postulating domination of higher brain/cortical regions over these systems might offer partial interpretation in this regard. Existence of a predefined programmed master regulatory scheme dictated by the life code embodied within the genome, and probably also within the proteome, of the cell might be a more comprehensive hypotheses. Unfortunately, currently available knowledge of the structure/function/behavior of the genetic material in unicellular, as well as in multicellular, organisms are far from being sufficient to offer plausible postulations as regards the nature, the location, the structure or the mechanisms of action of this hypothesized master life regulatory system.
The structural homogeneity of all components of the human genome rules out presence of any peculiar regions that can be alleged responsible for coding or defining such a master regulatory scheme. However, the marked functional heterogeneity of different components of the genome might suggest a provisional frame of such a master regulatory scheme. Within this context, three distinctive functional genome components that have crucial impact on genome size/genome structure/genome functions might participate in this presumed regulatory scheme. These include: 1. different classes of non-coding RNAs, short or microRNAs and long non-coding RNAs (lncRNAs). 2. different classes of the transposons. 3.Pyknons.
Non-coding RNAs exert crucial regulatory functions over genomic networks, and spontaneous or induced activities of transposons can affect genome size and homogeneity considerably particularly during development. Though very little is known about the functional significance of pyknons, their considerable sizable proportion in the genome and their peculiar structural features might suggest an important role played by pyknons as global regulators of gene function. They probably represent non-classic genes or non-classic transcriptional units capable of coding for, and regulating synthesis of, some microRNAs species for specific biological activities including regulation of genomic networks.
A plausible hypothesis regarding the nature of the postulated master scheme of life in living organisms is difficult to formulate in view of the marked paucity of current information concerning the detailed structural and functional organization of the genome, the different classes of RNA, and the proteome of living cells. Obviously, if a master dominating system responsible for defining the functional framework of the neural and endocrinal networks involved in regulating and mediating all life activities within the cell do exist, it must be located within, still functionally unidentified, regions of the brain. A marked different pattern of structural organization of the genome and RNA classes of cells constituting these master regions and a much wider spectrum of functional capabilities of their proteome might exist. Though current knowledge of the structures and functions of different regions of the human brain do not offer any helpful clues in this regard, future advances in molecular scanning techniques based on nano-scanners and nano-sensors capable of detecting the smallest, even the tiniest, structural deviations of the genome and the proteome, from currently known ordinary patterns, might reveal and disclose some of the mysterious aspects in this regard, and might even offer some answers to questions about the possible source and the exact nature of this postulated master scheme of life in living creatures.
5. Design of Biological Networks
The predefined designs of biological networks in living cells are dictated by information databases constituting a critical portion of the bioinformatics databases of life processes, probably contained within the genetic material of the cell. Unfortunately, the nature of these specific information databases defining all aspects of biological networks within specific cells or distinct organisms are ambiguous and largely unidentified. However, few rules can be delineated from accumulating observations in this regard. In general, biological networks conform, both structurally and functionally, to the general scheme of network systems though they have peculiar characteristics that differentiate them from other kinds of networks. Although all network systems are designed according to the functional requirements, biological networks are unique as they are formulated, constructed and regulated by preexisting programed designs contained within the cell/organism. The genome of the cell/organism comprises within its functional spectrum the databases needed for defining all network systems necessary for initiation/progression/preservation of life activities, as well as databases of networks responsible for inducing apoptosis, or programed termination, of these activities at certain stages of cellular life. The hierarchical design of life of all living organisms, with the exception of some viruses, has a more or less solid frame represented by the classic dogma of life at the molecular level which states that the genetic commands of the databases embodied within the structural/functional components of the genome are transcribed first to the transcriptome, then translated to the proteome that mediates all life activities of the cell/organism. All steps, and every step, of this design are under strict control of regulatory, proofreading, executive and corrective/repair networks composed of, and comprising all, structural/functional components of the cell. As processes and mechanisms of construction and maintenance of each structural component of the cell are mediated by network systems, nearly every cellular/subcellular component of the cell has its peculiar spectrum of networks mediating these mechanisms. The same applies to functional activities of the cell which are mediated and regulated by varying numbers of networks depending on the spectrum of cellular activities. The exceedingly large number of structural and functional biological networks in living cells function in concert, they are coordinated, interconnected and synchronized within a precise, punctilious context aiming at preserving the cardinal features of life at the molecular level as well as the whole organism level.
Biological networks have common routine functional features irrespective of the life activities mediated and regulated by these networks. Each network is composed of a varying number of structural proteins, catalytic enzymes, signaling molecules, and other organic/inorganic regulatory molecules necessary for network functions. The constituents of the network communicate with each other either directly for adjacent complexes, e.g., through structural conformational changes of proteins, or via signaling molecules that transport commands from regulatory units of the network, e.g., key proteins, to executive units, e.g., enzymes. Main products or secondary products of network function, e.g., ATP or metabolites, are either handled by other networks for specific physiological purposes or function as input substrates for renewal of network functions.
Although the major portion of the spectrum of biochemical and physiological regulatory networks and pathways through which the genome/transcriptome/proteome dominates all life activities at the molecular level inside the cell has been disclosed, and in spite of defining the major part of the spectrum of neural and endocrinal regulatory mechanisms of network functions on the whole organism level, the exact nature of the molecular mechanisms responsible for sensing and anticipating network demands by specific components, and the exact nature of the molecular mechanisms responsible for perception of these demands by executive components of the network remain extremely mysterious. Though occurrence of structural conformational changes in a protein, an enzymes or an organelle membranes upon contact, or interaction, with another biomolecule can be interpreted in terms of thermodynamics, and sometimes according to classic dynamics, occurrence of comprehensive organized and coordinated responses of all network components upon triggering/activating/initiating the first step in the pathway is hard to interpret by any currently available materialistic explanations.
6. Structural and Functional Distortion of Biological Networks
The predefined structural and functional design of biological networks aims primarily at ensuring optimal performance outcomes under variable working conditions. However, attaining and maintaining optimal functioning conditions of biological networks are dependent on presence of many factors and fulfillment of many conditions including: first: presence of properly constructed executive networks comprising all needed constituents, second: presence of intact regulatory networks or pathways necessary for controlling function(s) of executive networks, third: availability of properly adjusted continuous/interrupted flow of input substrates needed for initiation and keeping of network function(s), fourth: proper coordination with other functionally related networks or pathways responsible for handling output products of the network, fifth: properly synchronized renewal and replacement of network components, or biomolecules, that become structurally distorted and functionally inefficient over time.
Deviations from the proper predefined design of biological networks happen all the time throughout the whole life span of the living cell. These distortions are expected, or even inevitable, in view of the persistently dynamic nature of the network and the continuously changing circumstances of the cellular compartments enclosing the network(s). Underlying causes of these deviations can be deduced from analysis of factors participating in formation, regulation and disassembly of the network. Defects in the general framework design of the network affecting its mechanism of action might be inherited, e.g., congenital genetic disorders, or might be acquired following fertilization and zygote formation. Mutation-induced defects and abnormalities can present at any time and in any tissue/organ depending on the function(s) of affected networks. Similarly, mutations affecting genes or sets of genes responsible for synthesis of protein-enzyme components of the network or microRNA-protein regulatory factors have peculiar temporal and spatial profiles depending on the functions/localization of the network. Variations in environmental conditions of cellular compartments enclosing biological networks might be triggered and initiated by external factors like physical stresses, e.g., temperature changes, chemical stresses, e.g., changes in pH status, or biological stresses, e.g., viral or bacterial infections. These variations are dealt with, more or less efficiently, by networks specifically designed for these purposes as long as they occur within the predefined functional spectrum and the tolerable threshold limits of the network design. Over the threshold quantitative changes of these environmental variations, or exposure to external variations that can not be dealt with by the network, can result in a graded spectrum of functional deterioration of network performance starting with reduced outcome and ending with complete ravage, degradation or damage of the network. Exposure to ionizing radiations, for instance, leads to breakage of the sugar-phosphate backbone of DNA. Within limits, DNA repair systems or DNA repair networks can repair the resulting damage via the efficient DNA repair mechanisms comprising endonucleases/polymerases/ligases, among many other components of the repair system. Overexposure to lethal or intolerable doses of ionizing radiations, however, can cause widespread damage to DNA that can not be repaired, leading to irreversible devastating and drastic changes of the genome. The same scenario applies also to protein components of the networks, where exposure of living cells to lethal or damaging external stresses, like high temperature or irradiation, can lead to a wide range of detrimental consequences including damage of structural conformation of the proteins, disassembly and disorientation of the proper design of the different domains of the protein molecules, and loss of the external water envelope that constitutes an integral functional component of the molecule. Ensuing damage of the structural components of the network results in progressive failure in network performance, failure of physiological functions mediated by the network with consequent pathophysiological alterations and pathogenesis of pathognomonic disease depending on the functional spectrum of affected networks.
The cooperative and synchronized performance of functionally related sets or groups of networks, either locally in single cells or more globally in tissues/organs/organisms, is precisely controlled by, still unidentified, regulatory system(s) that ensures proper flow of input requirements or substrates to specific networks and parallel flow of metabolites or end products to other networks for final usage. Attaining and maintaining optimal life processes in living cells depend on the closed circle-like dynamic nature of this functional design of biological networks. Deviations from the predefined framework of this design or distortion of one or more of its constituent networks underlies the development of pathophysiological alterations and pathogenesis of disease. For instance, mutation-induced defective or deficient synthesis of components of an executive or input network will result in deficiency of products, biomolecules or ATP, necessary for functioning of other related networks or for provision of energy needed for performing, any and all, life processes in the cell. Similarly, mutation-induced defective or deficient synthesis of components of intermediary or output handling networks will lead to accumulation of products of executive networks and marked disturbances in the thermodynamic equilibria of involved reactions. Mutation-induced defective or deficient synthesis of structural components of regulatory networks, also, has a more widespread deleterious effects on all networks dependent on structural and functional integrity of these regulatory networks. The abovementioned examples reveal, in brief, some of the causes and effects of distortion of, or damage to, different types of biological networks and their role in pathogenesis of genetic as well as of non-genetic disorders consequent to these structural and functional alterations.
7. Mechanisms of Regulation of Biological Networks
Regulation of biological networks comprises nearly all functional aspects of the networks including initiation, activation, augmentation, inhibition, persistence, coordination and synchronization of function with related network groups, and termination of network function when required tasks are accomplished. Key networks might regulate function(s) of subsidiary networks through regulatory activator/terminator proteins, enzymes and microRNA. Inorganic components like heavy metals can also act as key regulatory molecules of some network functions 6. These regulatory proteins might exert their roles via acting as substrates or competitors of key enzymes of the network. Network activation depends on the type of the network, the nature of the regulatory network, the functional spectrum of the network, and the nature of the regulatory factors participating in defining network outcome. Metabolic networks are usually activated when the primary substrate is availed to the first initiating enzyme of the reaction cascade. Though all components of metabolic networks are synthesized under strict control of genomic regulatory networks, metabolic networks have a considerable degree of self-automation as long as the substrate resources and structural network components, enzymes/proteins/biomolecules, are available within the reaction environment. However, although core components of signaling networks, including signal receptors, are similarly synthesized under strict control of genomic regulatory networks, signaling networks do not have similar self-automation behavior like metabolic networks because they are primarily designed so as to elicit temporary and instantaneous regulatory responses. The second messenger outcomes of signaling pathways, also, do not have such degrees of self-automation because their effects are conditioned by many other factors like availability of transcription microRNA molecules, structural integrity of targeted genes, concomitant complementary and synchronized signaling pathways participating in the same task(s), and augmentation/inhibition by specific key regulatory molecules.
7.1. Regulatory Roles of Ribozymes and Riboswitches
Ribozymes, or ribonucleic acid enzymes, are RNA molecules capable of catalyzing specific biochemical reactions, similar to the action of protein enzymes. The most common activities of ribozymes are the cleavage and ligation of RNA and DNA, and peptide bond formation. Within the ribosome where translation and protein synthesis occurs, the functional part of the ribosome is fundamentally a ribozyme, composed of RNA tertiary structural motifs containing metal ions such as Magnesium as cofactors. Ribozymes within the ribosome structures function as part of the large subunit ribosomal RNA to link amino acids during protein synthesis. They also participate in a variety of RNA processing reactions, including RNA splicing, viral replication, and transfer RNA biosynthesis. Examples of ribozymes include the hammerhead ribozyme that catalyzes reversible cleavage and joining reactions at a specific site within an RNA molecule, the VS or the Varkud satellite (VS) ribozyme that carries out the cleavage of phosphodiester bonds, and the hairpin ribozyme present in some RNA viruses. Like metallo-enzymes, metal binding is critical to the function of some ribozymes, notably the leadzyme. The interactions induced and carried out by the ribozymes use both the phosphate group and the carbon core of the base of the nucleotide and result in major structural conformational changes of the molecule that seem necessary for its function (Figure 4 & Figure 5).
Figure 4.Structural design of ribozymes
Figure 5.Regulatory networks of riboswitches
Numerous mRNAs in prokaryotes carry complex folded domains, known as riboswitches within the non-coding portions of their polynucleotide chains. Each riboswitch directly binds a specific metabolite, without the obligate involvement of a protein factor, and then controls gene expression by harnessing changes in RNA structure to influence transcription elongation, translation initiation, or other aspects of the process that leads to protein production. In prokaryotes, most riboswitches are located in the 5'-UTRs of mRNAs, and are typically composed of two functional domains. The first is known as the aptamer domain encountered as the nascent mRNA that emerges from the RNA polymerase during transcription. The aptamer serves as a molecular sensor embedded within the riboswitch, where it selectively recognizes its corresponding target molecule within the complex sea of other metabolites. Like the majority of enzymes, each aptamer must selectively recognize a metabolite with the appropriate affinity. The second functional domain of the riboswitch is known as the expression platform located downstream from the aptamer domain, and is responsible for transducing metabolite-binding events into gene-control consequences by allosteric modulation of the structure of the 5'-UTR of mRNA 7. The discovery of metabolite-sensing riboswitches and other types of RNA sensors has revealed RNA-based mechanisms that cells use to regulate gene expression in response to internal and external changes. Structural studies have shown how these RNAs can carry out a range of functions. In addition, the contribution of ribozymes and riboswitches to gene expression is being revealed as far more widespread than was previously appreciated 8.
Although very few ribozyme categories have been characterized in eukaryotic cells, they probably play important regulatory roles essential to the integrity of the cell genome. For example, their ability to cleave RNA molecules imparts to the ribozymes an antisense down regulating function that might be used to regulate transcription defects due to overexpression of mutant genes in specific pathological states, e.g., oncogenes in malignant cells. Within this context, they might be considered as part of the anti-mutation mechanism system of the cell that protects the stability and integrity of its genome. As possible potential regulators of many aspects of gene expression, hypotheses postulating the participation of ribozymes in construction of genomic regulatory networks, as down regulatory units, seem quite reasonable.
7.2. Regulation of Higher Genomic Regulatory Networks
The crucial roles played by the genome in defining the proteome, and the formulation of the exceedingly large number of networks composed of proteome components, poses many puzzling questions regarding the nature and the mechanisms responsible for regulating the functions of the genome itself. The adopted dogma of molecular biology reflects the current interpretation of life processes at the gene level, which represents the final level amenable to observation, analysis and interpretation of life processes in living cells. The hierarchical design of the biological master system responsible for controlling the flow of regulatory information and executive commands from the genome to the networks is apparently reasonable and clearly understood to a large extent. The nature of this master system, however, remains totally mysterious and completely elusive as regards the sources and the mechanisms of action of all its aspects. This dilemma probably represents the real challenge facing trials to define life within the context of molecular biology. In spite of the tremendous information gathered during the relatively long time since the beginning of the debates and assumptions regarding the nature or secret of biological life, and in defiance of the parallel tremendous advances in establishing diagnostic techniques capable of revealing the nature and behavior of the biomolecules and the atoms at their nano-scale, no reasonable or acceptable hypothesis as regards the source, the location or the nature of master system that defines biological life has yet been presented. Hypotheses attributing beginning and persistence of life to chance occurrence and automation are futilitarian injudicious postulations that are not worthy of any logical consideration. Though the alternative postulation which adopts the religious interpretation of the creation, the beginning and persistence of life can not be debated or discussed within a scientific context being dependent solely on metaphysical and supernatural assumptions that are not amenable to scientific interpretation, it probably represents a more logical and realistic alternative, at least, until undisputed facts regarding the nature of life can be presented. Away from ignoring and avoiding nonsense debates and useless discussions about occult aspects of life, adopting this religious alternative will help in saving money and efforts for useful researches aiming at improving our understanding of the mechanisms underlying development of human diseases and the best prophylactic and therapeutic approaches to avoid and combat these diseases.
8.Types and Classification of Biological Networks
Classification of biological networks is difficult and perplexing in view of the exceedingly large numbers of primary/central/pivotal key networks and corresponding, even larger, numbers of secondary/subsidiary/ temporary networks present in each living cell. Additionally, the wide spectrum of structural and functional characteristics that distinguish each of these network categories and subcategories makes their comprehensive classification an arduous task. Many cellular networks function in concert within a common and shared context imposing functional complementation to mediate specific tasks within a larger network system, e.g., cell cycle regulating network. Moreover, functional complementation of networks may comprise different network categories, e.g., transcription regulatory networks and signal transducing networks, or similar category networks working in succession in a cascade manner, e.g., metabolic networks.
No single reasonable and acceptable classification of biological networks could be formulated because of the many different parameters relied upon for classification. For example, classification may depend on network function, e.g., regulatory/developmental/executive networks, or on structural components of the network, e.g., protein-enzyme networks, protein-microRNA networks and mixed network comprising other components like inorganic signaling molecules. Other parameters of classification schemes include complexity of the network, e.g. major/subsidiary/intermediary networks, and cellular location of the network, e.g., nuclear/mitochondrial/ cytoplasmic networks. Each of these network categories could be classified, further, into many subtypes according to peculiar characteristic features of the network. Overlapping and interlacement of network subtypes is common because many networks share common features with other apparently distinctive network types. For example, metabolic networks could be classified according to their structural components as protein-enzyme networks and as executive networks based on their functional specialization. Some metabolic networks function as major pivotal or central networks while other less important metabolic pathways serve as subsidiary networks. In addition, metabolic networks could be, also, classified as cytoplasmic, mitochondrial or nuclear metabolic networks according to their location within the cell compartments.
Another classification scheme of biological networks worthy of consideration is based on the temporal nature and persistence of network function. According to this classification scheme, four main categories of biological networks could be identified with clear discrimination between the peculiar characteristics of each of them: 1. Continual unceasing networks functioning all through the life span of the cell, e.g., energy producing networks and cell division regulatory networks. 2. Temporary networks that function during particular stages of the life span of the cell, e.g., developmental networks. 3. Circadian rhythm networks implicated in regulating physiological activities characterized by having nighttime/daytime cyclic endogenous physiological rhythms, e.g., sleep/wakefulness, blood pressure fluctuations, rhythmic secretion of hormones/endorphins, and entrainment or synchronization of these activities with environmental cycles of light and dark. 4. Oscillatory rhythm networks that work in an interrupted recurring pattern in response to external environmental changes, e.g., temperature cycles, or in retroaction to internal changes in cellular homeostasis, e.g., electrolyte balance. However, functional classification of biological networks seems to be the most reasonable as functional specificity of the network is the main determinant of its importance within the life frame of the cell [Table 1].
Table 1. Types and classification of biological networks| Type of network | Examples | ||
|---|---|---|---|
| 1. According to hierarchical organization | 1. Master networks | ||
| Networks responsible for maintaining identity, integrity and stability of the genome. | |||
| 2. Intermediary networks | |||
| Networks responsible for synthesis of the proteome. | |||
| 3. Executive networks | |||
| Metabolic networks responsible for direct mediation of life activities. | |||
| 2. According to function | 1. Regulatory networks | ||
| A. Genomic regulatory networks | |||
| 1. DNA replication networks | |||
| 2. Replication proofreading networks | |||
| 3. DNA repair networks | |||
| B. Transcriptional networks | |||
| a. Post-transcription modification networks | |||
| b. mRNA editing networks | |||
| C. Translation regulatory networks | |||
| a. Ribosomal networks | |||
| b. mRNA recycling/degradation networks | |||
| D. Signal transduction networks | |||
| a. Wnt signaling pathways | |||
| b. Hedgehog signaling pathway | |||
| c. Notch signaling pathway | |||
| d. Apoptosis signaling pathway | |||
| 2. Developmental networks | |||
| 3. Executive networks | |||
| a. Metabolic networks | |||
| b. Transport networks | |||
| c. Post-translation modification networks | |||
| d. Apoptosis networks. | |||
| e. Cell cytoskeleton/membranes biogenesis networks | |||
| f. Oxidative-phosphorylation networks | |||
| g. Xenobiotics intoxication/disposal networks. | |||
| 3. According to structural components | 1. Gene networks | ||
| 2. Protein-enzyme networks | |||
| a. Metabolic networks | |||
| b. cytoskeleton networks | |||
| 3. Non-coding microRNA/lncRNA networks | |||
| 4. According to cellular location | 1. Nuclear networks | ||
| 2. Mitochondrial networks | |||
| 3. Cytoplasmic networks | |||
| 4. Specific cellular organelles networks | |||
| 5. Extracellular networks | |||
| 5. According to persistence of function | 1. Continual unceasing networks | ||
| a. Energy production networks | |||
| b. Cell cycle networks | |||
| c. Vital neurogenic networks | |||
| 2. Temporary networks | |||
| a. Developmental networks | |||
| b. Stress networks | |||
| 3. Oscillatory rhythm networks | |||
| a. Circadian rhythm networks | |||
| 1. Sleep/wakefulness networks | |||
| 2. Blood pressure regulatory networks | |||
| 3. Melatonin/hormones/endorphins secretory networks | |||
| b. Intrinsic rhythm networks | |||
| 6. Neuronal networks | 1. Recognition/reception/processing/responding to stimuli networks. | ||
| 2. Storage/processing/retrieval of information networks. | |||
| 7. Other networks | Networks of theoretical interests | ||
| 1. Intra-species (within-species) interaction networks | |||
| 2. Inter-species (between-species) interaction networks | |||
| 3. Food-web (feeding interactions) networks | |||
9. Temporal Classification of Biological Networks
The aforementioned resume of different network categories in the living cell outlines, in brief, what could be referred to as the dynamics of life processes in living cells cell. Analysis of the classic dogma of life cycle, traditionally and illogically referred to as the classic dogma of molecular biology, reveals the three major structural components responsible for mediating life activities in living creatures, i.e. the genome, the transcriptome and the proteome. It also defines the major network categories which guarantee precise and optimal performance of these activities. Additionally, it delineates the temporal relations between the biological implications and the dynamic cascades of these vital life activities. Within this context, a time-dependent or time-related temporal classification of biological networks might be assumed to comprise the following postulated network categories hierarchically arranged according to the temporal cascades of their functions:
A. Crucial Stage or Genome Preserving Network Category composed of many different types of networks responsible for maintaining the identity, the integrity and the stability of the genome. This life sustaining and maintaining obligation starts at fertilization and continues till cell death occurs.
B. Transcription Stage Network Category also composed of many different types of networks that regulate all aspects of transcription including initiation/enhancing/silencing and termination of transcription, post-transcription modifications and availing mRNA to protein synthesis machinery in the cytoplasm.
C. Translation Stage Network Category composed of regulatory and executive networks responsible for controlling translation. Translation of mRNA coding for proteins results in synthesis of the exceedingly large number of structural and catalytic proteins which compose the proteome. The proteome provides the structural protein components of all the biological networks responsible for direct execution and mediation of all life activities of the cell.
D. Executive Stage Network Category which comprises all types of networks and pathways directly involved in regulation and performance of all life activities in the cell. This category includes the vast majority and the largest portion of biological networks in the cell.
E. Final Stage Network Category including networks responsible for initiation and completion of apoptosis of senescent and heavily mutated cells. Apoptosis networks and pathways are a subcategory of executive biological networks that finalize the life cycle of the cell and put an end to the biological existence at the cellular level.
10. Functional Classification of Biological Networks
Functional classification of biological networks, probably, represents the most plausible approach in this regard. As referred to earlier, the classic dogma of life cycle denotes the presence of three major structural and functional components in the living cell responsible for controlling all life activities of the cell, viz. the genome, the transcriptome and the proteome. Consequently, three main network categories in the cell could be identified and delineated comprising genomic, transcriptomic and proteomic networks. Analysis of the nature, the composition, the dynamics and the functions of each of these three main network categories reveals that each category has distinctive characteristic features defined, primarily, by their basic functions. Functional characterization of each of these three basic network categories and the subsidiary networks and pathways dependent on, and regulated by, each of them discloses, with very few exceptions, the global framework of life processes in living cells. Presence of secondary/subsidiary/tributary and intermediary networks tightly connected and properly synchronized to ensure optimal performance of their functions is mandatory in view of the complexity and multistage nature of life processes in the cell. However, it must be emphasized that this classification is speculative and is meant for educational purposes in the first place. In fact, sharp demarcations between the individual spectra of each of these categories is practically, and even theoretically, impossible because of the intimate and cooperative relationships between their functional spectra which are widely intermingled and complementary (Figure 6, Figure 7, Figure 8).
Figure 6.Functional Classification of Genomic Regulatory Networks
10.1 Genomic Networks
There is no globally accepted definition or agreed upon classification scheme of genomic networks. This is attributed to the confusing overlap between the terms genetic/genic/genomic as regards their qualitative profiles as well as their quantitative spectra. Additional confusion is met with upon trying to demarcate the imbricate and mutual relationships between the genome and the proteome in construction and organization of the network, since carrying out and execution of functions of all biological networks, irrespective of the type or the nature of the network, are totally dependent upon proteome components, viz. proteins/enzymes/small peptides etc. Furthermore, the crucial roles played by signaling pathways in initiating, regulating and modifying the performance of all types of networks, including genomic regulatory networks, adds more difficulties to theoretical postulations aiming at formulate an accepted comprehensive framework as regards the nature of genomic networks. However, a preliminary tentative definition approach could define genomic networks as a network system primarily responsible formaintaining the identity, the integrity and the stability of the genome of the cell. Genomic networksare functionally continual unceasing networks located mainly within the cell nucleus. Though their continuous unceasing functions begin with fertilization and zygote formation and do not stop until biological death of cellular functions occurs, they represent actual continuation of the functions of ancestor networks performing the same genome preserving functions in the ova and sperms, that participate in fertilization.
Genomic regulatory networks might be tentatively classified into many functional categories including networks composed of master genes responsible for preserving the identity/integrity/stability of the genome, networks composed of regulatory genes responsible for regulating the functions of structural genes, networks composed of structural genes responsible for controlling protein synthesis in the cell, and transcriptome networks responsible for regulating and synchronizing the varying functions of different RNA categories in the cell (Figure 6).
Although the structural design of genomic networks resembles the general design of most other biological networks in being constructed mainly from proteome constituents, viz. proteins and enzymes, a characteristic feature in this respect is inclusion of many different classes of microRNAs within this design. Small or microRNA molecules constitute critical regulatory components of most types of genomic regulatory networks. Most genomic networks are composed of, and include sets of functionally related genes, temporally and functionally organized and interconnected in a synchronized hierarchical pattern, to perform specific shared functions. Genomic networks comprise large numbers of vital and important groups of secondary networks with varying functions including, for instance, networks responsible for controlling and regulating DNA replication, replication proofreading and DNA repair, DNA-associated proteins, cell cycle initiation and cell division. Other, structurally and functionally, still, undefined and largely speculative networks probably exist and exert indispensable tasks within the global context of genome preservation and behavior. For instance, it seems reasonable to assume that there are subsidiary or intermediate genomic networks that act as sensory or receptive pathways responsible for detecting the need of the cell to multiply, others that detect the need of the cell to the product(s) of specific gene/cluster of genes, and other pathways that trigger activation and transcription of target genes and regulate the entire process of gene function until proper gene products are synthesized, modified, trafficked, properly located and become ready for participation in physiological activities in the cell.
Although genomic networks regulate all life aspects in the living cell, precise details concerning the origin of their design, the mechanisms of their formation, and the nature of the master system(s) that dictates the commands and rules controlling and regulating their functions, are totally unclear. This reflects the eternal ongoing dilemma of biology, and science in general, where we can observe how things happen and analyze their outcomes which reveal why they happen, but we remain totally benighted of the basic dilemma: how things exist from the start.
10.1. Genome Preserving Networks
This group of genomic networks are responsible for guarding and maintaining the three basic features of the genome, viz. genome identity, genome integrity and genome stability, which represent the vital core and the essential framework of life of living organisms. These three distinct fundamental aspects of the genome constitute the bases for keeping biological diversity of living organisms and ensuring the biological fitness necessary for reproduction, proliferation and persistence of kind of each organism. Guarding genome identity guarantees the persistence of species-specific characteristic features of the organism. This task is accomplished through strict and precise regulation of proper replication of DNA and proper duplication of the entire set of chromosomes of the dividing cell. Normal cell division and formation of daughter cells with identical genomes ensures preservation of the genetic identity of the cell and the organism. Conserving genome integrity represents a crucial function since maintaining normal life processes in the cell depends on the cooperative and synchronized functional participation of all structural constituents of the genome. Loss or non-repairable damage of parts of the genome, e.g., because of mutagenic events or due to exposure to non-mutagenic external or environmental stresses, results in consequent loss of functions of lost or damaged regions. In the vast majority of these instances, affection of physiological functions regulated by lost or damaged regions of the genome occurs and may result in a wide spectrum of pathological alterations, depending on the qualitative and the quantitative extent of the lost functions. Drastic or detrimental consequences of affection of genome integrity include, for example, temporary reduction in efficiency of cellular activities, progressive failure of cellular functions, development of tumors/immunodeficiency/neuro-deterioration or other disease conditions in humans and other susceptible creatures, development of congenital malformations consequent to disruption of genome integrity during development, and final cell death. Preserving genome stability represents a third crucial goal of genomic regulatory networks. Precise functioning of all regions of the genome necessitates stabilization of its components. This stabilization involves the spatial relations between different genome components, e.g., the gene locus concept or the predefined locations of genes and non-genic segments of the genome over specific chromosome regions. This essential spatial design of genome components that preserves genome stabilization can be disturbed through different pathogenetic mechanisms including mutation-induced chromosomal rearrangements, spontaneous and/or induced activation of transposons, and insertional mutagenesis caused by some viral infections. DNA and chromosomal repair mechanisms and suppression of transposon over activity, e.g., by piwiRNA particularly during early development, represent important anti-mutation mechanisms mediated by genome stabilizing networks to keep and preserve genome stabilization, and normal cellular activities, all through the life cycle of the living cell and the whole organism.
Preservation of the mitochondrial genome does not deserve much efforts like the nuclear genome in view of the multi-copy nature of mitDNA. Preservation of nuclear genome is critical as hemizygosity and/or heterozygosity of nuclear genotypes can result in a wide range of pathological alterations. The occurrence of large numbers of multiple copies of the mitochondrial genome in the cell ensures continuation of physiological functions dependent on mitochondrial genes even if considerable loss of a portion of these genes happens. This might, partly, explains the absence of some repair mechanisms for mitDNA in contrast to their presence for repair of nuclear DNA defects.
Though the exact nature of the structural designs and the functional spectra of genome preserving networks are not yet disclosed, networks implicated in, and responsible for, genome preservation would probably include a wide variety of networks includinf, for example, networks sensing the need for DNA replication and ensuring proper initiation and completion of the whole replication process, networks responsible for replication proofreading including repair of DNA mutations and proper organization of DNA-chromatin components, networks regulating expression of gene families coding microRNAs classes involved, either singly or as parts of more complex pathways, in performing critical regulatory tasks, networks regulating programmed targeted activities of transposons as well as networks responsible for repair of genomic mutations induced by spontaneous activities of transposons via e.g., synthesis of piwiRNAs classes and possibly through other mechanisms, networks that regulate the dynamics and different aspects of the cell cycle and cell division, networks regulating the formulation and functions of the considerable large number of developmental networks that start their functions at fertilization, networks regulating apoptosis, and many other tributary networks.
Inclusion of apoptosis regulatory networks as subsidiary genome preserving networks seems reasonable in view of the important biological functions performed by apoptosis of heavily mutated and damaged genomes. Within a biological context, apoptosis of mutated genomes saves the cell the drastic fate of malignant transformation which ends and leads, in the majority of instances, to death and extinction of the whole organism. It also saves the cell the detrimental fate of progressive deterioration and loss of cellular functions with ultimate loss of biological fitness and death. Apoptosis of heavily mutated genomes have an important and critical role in avoiding and preventing the emergence of mutant phenotypes not conforming with the optimal standards of the classic dogma of life cycle. Multiplication of mutant phenotypes would, probably, add more aberrant mutant genotypes to the genome pool of affected populations leading, ultimately, to either interruption or final disruption of the normal life cycle of affected organisms.
10.2. Transcription Regulatory Networks or Transcriptome Networks
Transcription regulatory networks comprise large numbers of essential tributary, subsidiary and intermediary network groups in view of the indispensable roles played by the transcriptome in nearly all crucial cellular functions including regulation of gene functions and, even, modulation of many aspects of genome stability and behavior. The considerable large numbers and varying types of RNA classes in living cells imparts critical significance to, and reflect, the pivotal functions of the transcription regulatory networks. It is estimated that the human genome probably encodes 1,400 transcription factors that regulate the expression of more than 20,000 human genes 9. In addition to messenger or mRNA which, probably, constitutes the major portion of the transcription products in the cell, the transcriptome comprises many other functional categories of RNAs indispensable for regulating all life processes in the cell, including ribosomal or rRNA, transfer or tRNA, editing or guide RNA, small or microRNA, long non-coding or lncRNA, PIWI-interacting or piRNA, in addition to many other, still undefined, classes. The transcriptional network(s) responsible for regulating the process of transcription requires, also, many additional subsidiary networks to render the primary heterogeneous transcribed mRNA molecule ready for performing its functions in the cytoplasm. Each of the post-transcription modification processes of the primary nuclear mRNA, e.g., capping/polyadenylation/ circularization/excision of introns/splicing of exons/editing and transport across the nuclear membrane, is mediated by distinct functional network(s) composed of peculiar sets of proteins and enzymes and other factors working with other networks in a strict interconnected and synchronized pattern to guarantee proper accomplishment of the final goals of transcription (Figure 7,Figure 8, Figure 9).
Figure 7.Regulatory networks involved in transcription
Figure 8.Regulatory networks involved in protein synthesis
10.3.Translation Regulatory Networks or Proteome Networks
Translation networks are responsible for synthesis of the proteome, or the sum total of the specific set of proteins coded by a specific genome in the living cell. In addition, they are responsible for defining and regulating the different aspects of the proteome. These aspects include types and amounts of synthesized proteins and the rates of their synthesis, structural post-translation modifications of synthesized proteins and enzymes, and protein trafficking and localization within intra and extra cellular locations. Translation networks responsible for synthesis of protein products coded by structural genes comprise large numbers of subsidiary networks each concerned with performing a particular step of the translation process. Pre-translation networks, including specific metabolic regulatory networks that ensure availing sufficient amounts of required amino acids in the cytosol, have a crucial role in the pre-translation process. Their postulated existence is supported by the finding that amino acid supply positively correlates with translation efficiency and ribosome density, and that metabolic amino acid supply facilitates ribosome utilization, possibly by buffering ribosome density change against amino acid starvation, thus helping to maintain a relatively stable translation environment 10. Other postulated pre-translation networks might include networks responsible for ensuring availability of the exceedingly large numbers of different factors needed for the translation process. The ribosomal networks, then, begin decoding the mRNA in conjunction with the tRNA networks that get the proper amino acids into the translation site in the cytoplasm and a third metabolic network, responsible for elongation of the newly synthesized polypeptide chain, works until the proper length of the chain has been reached. After synthesis of the proper polypeptide chain, protein-protein interaction networks begin their roles in inducing post-translation structural modifications of the synthesized protein, e.g., folding and binding to other proteins, to render it structurally competent for performing its physiological functions. A final set of networks then take the responsibility of regulating post-translation trafficking and localization of the synthesized proteins at their proper destinations whether inside the cell, at the cell membrane, in the extracellular spaces or at other distant sites in multicellular organisms (Figure 10, Figure 11).
Figure 10.Transcription-translation regulatory networks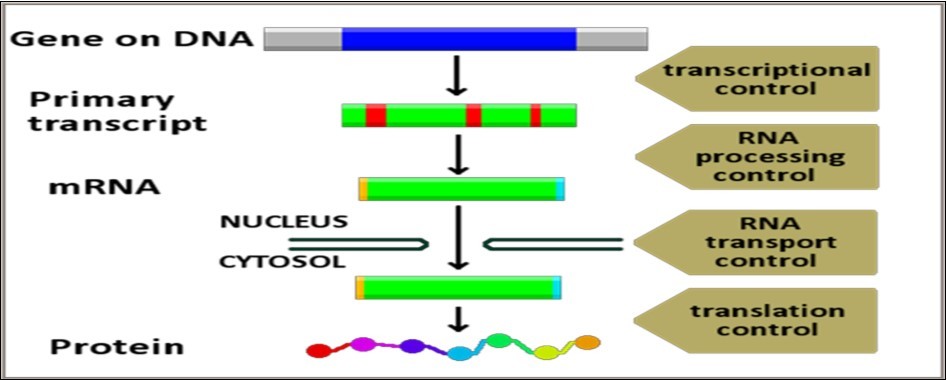
10.4. Developmental Networks
Development is a very complicated multistage biological process that requires utmost precise coordination of all cellular activities occurring in the zygote as well as in all descendent daughter cells. It also necessitates optimal temporal synchronization of activities of all regulatory/signaling/metabolic networks that control all aspects of development. Development begins by fertilization and zygote formation and reaches its end by giving birth to a fully developed viable fetus. Though this definition is literally accepted within the context of developmental genetics, it does not reflect the fact that development is a continuous process that persists all through the life span of the organism. However, according to this traditional concept, development comprises three major distinctive stages: 1. Fertilization and early post-fertilization genomic rearrangements and alterations, 2. Cell growth, cell division and multiplication, differentiation, specialization, localization, migration and apoptosis. 3. Morphogenesis of specialized tissues (histogenesis) and organs (organogenesis) in accordance with the genetically determined programmed and predefined species-specific pattern and position of all anatomical parts and landmarks of the developing embryo. Embryonic development in mammals, or embryogenesis, in turn progresses along four distinctive consequent stages: cleavage, patterning, differentiation and growth. Each of these stages involves many structural/functional dynamic alterations of nearly every group of cells participating in performing specific function(s) during development.
Developmental networks that form/appear at fertilization to regulate embryogenesis and fetal development have peculiar features, some of which are bewildering. They are formulated and structured de novo following fertilization and begin working in addition to the already existing networks of the zygote. Their very early appearance, probably, reflects the reprogramming processes that characterize the genome of the zygote that behaves in a distinctive manner different from that of the ovum and the sperm. The functions of developmental networks continue until giving birth to, or completing the formation of, the newly created organism. Some specific developmental networks, however, continue functioning even after birth because many fetal functions are not yet fully developed and become mature only after birth during the post-natal life, sometimes even decades after birth. Developmental networks comprise many categories of temporary networks and pathways that work only during these crucial periods of life. Though most of these temporary networks become functionally concealed or structurally disassembled after performing their roles in development, many of their component proteins continue to function after birth. Proteins and enzymes that have key regulatory or signaling roles in cellular activities continue to function for life. However, the reappearance of many of these temporary developmental networks at later stages of life, sometimes decades after birth, in some pathological conditions, for example, as part of the pathophysiological spectrum of the malignant phenotype of many tumors, has no clear-cut explanation.
10.4.1. Characteristics and Regulation of Developmental Networks
Developmental networks have some peculiar features as regards mechanisms of regulation of their formation, their functions and their fates.
First: Similar to metabolic networks, all structural components of developmental networks are synthesized under strict control of genomic regulatory networks. However, contrary to the self-automation behavior of metabolic networks, meticulous control and persistent regulation of all functional aspects of developmental networks continues round the clock all through all stages of development. This is expected in view of the intended long-lasting goals of these networks aiming at ensuring complete and proper development of a normal shaped healthy organism. Precise coordination and synchronization of functions of the huge number of developmental networks acting during a limited, relatively short, period of life represent, probably, one of the most important and crucial transcendental life preserving and species-specific conserving aspects of life of all living creatures. Developmental processes including cleavage, gastrulation, patterning, differentiation, speciation, growth and apoptosis are basic indispensable processes that have to occur at particular sites during particular stages of development. Genetically determined defects or non-genetic teratogenic effects leading to faulty regulation with consequent disordered initiation/continuation/termination of any of these vital processes during development can result in development of a countless number of congenital defects and malformations.
Second: Though master genomic regulatory networks define and control the spatial and temporal frameworks of developmental networks, many locally-functioning signaling networks and modulator pathways play crucial regulatory roles over developmental processes during all stages of development. Additionally, local subsidiary metabolic/signaling regulatory networks and pathways are temporarily formed and constructed during specific stages of development in specific regions of the developing organism to perform particular tasks necessary for proper and intact development and growth of the newly created organism. After completion of their tasks, most of the temporarily formed developmental networks and pathways are degraded and disassembled.
Third: In addition to precise spatial coordination and synchronization of developmental cellular processes in particular regions of the developing organism, i.e. cell groups/tissues/organs, concomitant temporal coordination of these processes is indispensable for normal development. Though the exact details of the mechanisms underlying temporal regulation of developmental processes are mysterious and far from being fully comprehended, two regulatory mechanisms, each mediated by a specific network category, seem to play important roles in this regard. Oscillatory rhythm networks and signaling networks seem to be the main regulatory systems responsible for temporal coordination of developmental processes in humans, and probably other organisms. Contrary to circadian rhythm networks that coordinate physiological functions with daily changes in the day-night cycle, developmental oscillatory rhythm networks, most likely, do not work in accordance to the day-night cycle pattern of adult organisms regulated, for the most part, by Melatonin. The reason for this postulation stems from the finding that melatonin is not synthesized in embryonic/fetal tissues as the pineal gland becomes mature only after birth. Although melatonin receptors are widely expressed in embryonic and fetal tissues since early stages of development, maternal melatonin transplacentally delivered to the embryo/fetus represents the sole source of this hormone during development. Important temporal regulatory roles, and neuro-protective functions, of melatonin during development have been suggested 11, nevertheless, mechanisms of temporal regulation of developmental processes by melatonin are quite obscure. It is hard to conceive a cause and effect relationship between changes in the day-night cycle of pregnant mother and temporal coordination of ongoing developmental processes in the fetus. It might be more reasonable to suggest presence of specific developmental oscillatory rhythm networks driven by temporal changes resulting from the peculiar courses of local biochemical and biophysical alterations occurring in developmental regions. Thus, precise temporal coordination of developmental processes might be conceived as a cascade of ordered responses to succession of events/alterations rather than to succession of time periods related to the day-night cycle circadian rhythm.
Fourth: The possibility of prenatal or postnatal reformation of any of the developmental networks and pathways as repair mechanisms in case of occurrence of structural defects or loss of already formed tissues or organs has to be considered in view of the ability of some living creatures to regrow lost parts of their organs. Many creatures show, even, an amazing potential for total regrowth from just a piece of their bodies, e.g., cut pieces of the flat worms Planarians of the Turbellaria class, can grow into new full worms. This mysterious ability of reformation and regrowth of lost parts or even a whole new organism following trauma or autotomy, or self amputation, is possessed by many and widely varying animal species including spiders, reptiles, amphibians, sea cucumbers, sharks, starfish and many others 12. However, the recent observation of occurrence of this, not fully yet understood, phenomenon in mammals, where at least two species of African spiny mice, Acomyskempi and Acomyspercivali, were shown to be capable of completely regenerating damaged skin tissue through regrowing hair follicles, skin, sweat glands, fur and cartilage with little or no scarring 13. This potential intrinsic regenerative ability of some species to regrow lost parts of their bodies is different from organ regeneration processes which happen in many human organs e.g., thymus, adrenal gland, thyroid gland, intestine, lungs, heart, liver, blood vessels, germ cells, nervous system, eye tissues, hair cells, kidney and bladder, skin, hair follicles, pancreas, bone, and cartilage after partial loss of their parts, which represents replacement of lost parts by newly formed and multiplying cells of the remaining intact tissue 14.
Though no acceptable explanations of the nature or the mechanisms of the potential of regrowth of lost parts of some organisms are available, few assumptions regarding its possible underlying mechanisms might be offered based on occurrence of similar responses to local damage of tissues or cell masses during development. Necessity to compensate for lost function(s) might urge the reformation of the regulatory and executive developmental networks responsible for histogenesis and organogenesis of lost parts. Also, the preservation of the structural integrity and the anatomical profile of the organism represents a crucial aim of utmost importance during normal development. Loss of vital or important structural components of the organism that results in distortion of its anatomical conformation is a hazardous event for the developing organisms that may account for the reformation of developmental networks. Many factors might be assumed to participate in providing the suitable micro-environment(s) required for this developmental reformation process including the predefined structural design of the organism, the effects of intercellular connections and extracellular interactions, the predefined hierarchical sequencing of the structural and functional roles of the lost part within the whole context of development, and many others. However, the question of whether these postulated causes, which are observed during prenatal developmental processes, might also apply for similar situations of organ loss in post-natal life is, still, far from being settled for sure.
10.4.2. Developmental Signaling Pathways
Developmental signaling pathways constitute, and work within the frame of, key pivotal regulatory networks responsible for regulation, coordination and synchronization of the innumerable sequential and simultaneous stages and processes of development. Their roles are kept in precise harmony within their functional spectrum and within the wider global spectrum of cellular activities by, still, unknown mechanisms. Disturbed network function(s), due to genetic mutations leading to synthesis of defective protein components, has been causally linked with a wide range of teratogenic developmental defects that can involve nearly every tissue or organ. Meticulous analysis of network components and mechanism(s) of action during development would, surely, has a major impact on researches aiming at uncovering pathogenetic mechanisms that underlie pathogenesis of teratogenic malformations. Hopefully, prophylactic intervention, e.g., supply or replacement of defective network protein/enzyme components during pregnancy, might offer a possible preventive measure of many types of congenital malformations.
Signal transduction pathways are crucial in the regulation and coordination of all stages of embryonic and fetal development. Developmental signaling networks comprise mainly the wingless related (Wnt) pathways, the hedgehog (HH) pathways, the notch pathways, and the fibroblast growth factor signaling pathway. Additional subsidiary pathways include transforming growth factor-β (TGF-β) pathways, receptor tyrosine kinase (RTK) pathway, Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathways, and nuclear receptor pathways. These pathways have diverse structural components and complex biochemical mechanisms. However, they have in common a crucial function, viz. the activation of specific genes through the regulation of signal-dependent transcription factors. These target genes code for proteins/enzymes/RNAs required for the mediation and regulation of different stages of development either via exerting direct effects on key processes or reactions of developmental networks or by participation as structural components of these networks.
The functional significance of developmental signaling pathways covers a wide spectrum of vital processes necessary for normal development. These pathways work under strict control of regulatory networks in finely tuned concert characterized by many peculiar features including sharing of many pathways in performing certain developmental processes, mutual augmentation/inhibition of function(s) of some pathways by other pathways, and functional specificity of certain individual pathways in conduction of specific developmental processes. Main developmental signaling pathways include:
1. The Wnt signaling pathways which plays critical roles in defining body axis patterning, cell differentiation and specification, cell proliferation, and cell migration. These processes are necessary for proper formation of important embryonic and fetal tissues including bone, heart, and muscle. Increasing glucose uptake from the bloodstream in skeletal muscles has been found to be partially mediated by activation of Wnt/β-catenin signaling which enhances cell's sensitivity to insulin. This effect has a significant impact on the developing fetus in view of the major role of insulin in regulating and enhancing fetal growth.
2. The Hedgehog signaling pathway constitutes a critical signaling regulatory network during development where it regulates growth, patterning and morphogenesis of many tissues and organs. Disruption of Hedgehog signaling mechanisms during embryonic development due to mutations or teratogenesis has been causally linked to a wide spectrum of severe developmental disorders, e.g., holoprosencephaly.
3. The Notch signaling pathway is important for cell-cell communication required for cell differentiation processes during embryonic life. This pathway, also, has important regulatory roles in many early and late developmental processes of most vital tissues and organs including the heart, the brain, the pancreas, the mammary gland, the blood vessels, the immune system, and many others.
4. The fibroblast growth factor signaling pathway (FGFs) has pivotal regulatory roles during development. It comprises a large number of multifunctional extracellular signaling proteins and peptides that participate in many developmental processes including antero-posterior axis patterning, limb development, angiogenesis, and neural development 15.
10.5. Signal Transduction Networks
Precise regulation and maintenance of life processes in living cells necessitates proper coordination and optimal synchronization between all components involved in these activities. These components include central determinant systems that comprise the genome/the transcriptome/the proteome, structurally organized components that comprise all cellular organelles and extracellular sites where these processes take place, regulatory and executive systems that perform their functions through the different kinds of networks which directly mediate all life processes in living organisms (Figure 12 & Figure 13).
Figure 12.Concept of signal transduction
Figure 13.Mechanism of signal transduction
Optimal coordination and synchronization of life processes in living cells is achieved by highly organized and complicated system of communications between cellular components performing these processes. This communication system, referred to as cell signaling pathway, consists of large numbers of signaling networks or signaling pathways, composed of proteins/enzymes/small molecules. Cell signaling is a central regulatory mechanism that participates in all life processes in living organisms all through their life span, starting with fertilization and ending with apoptosis. Cell signaling networks constitute a pivotal life keeping component that regulates and coordinates intra-cellular activities as well as inter-cellular actions and cellular responses to the extracellular environment. Signaling networks exert their functions via different mechanisms through varying types of signaling pathways, each pathway representing an ordered sequence of reactions elicited and initiated by signal molecules, mostly proteins, which activate receptor proteins leading to a biochemical and/or biophysical change in specific components targeted by the pathway 16.
Signal transduction refers to the process of inducing specific change in receptor molecules by inciting signal molecules. The induced biochemical/biophysical changes of targeted components, or transducers, result in initiation and/or termination of function of specific networks, e.g., genic/metabolic/developmental/regulatory networks. Many signaling pathways result in signal transduction cascades leading to activation of specific transcription factors and consequent activation or suppression of specific genes with resulting increase or decrease in synthesis of products of targeted genes, respectively. These products include structural proteins, catalytic enzymes and regulatory RNAs. Consequent to these changes, particular physiological processes regulated by networks structured and/or regulated by these products are initiated, maintained, or terminated. As cell signaling mechanisms control spatial and temporal cell perceptions and responses to changes in intracellular as well as extracellular microenvironment, errors in cell signaling system are expected to result in causation and development of a very large number of genetic and non-genetic disorders including, for example, developmental malformations, diabetes mellitus, inborn errors of metabolism, autoimmnue disorders, cancer, and neurodegeneration, 17, 18, 19.
Mechanisms of intracellular and intercellular communications comprise two apparently different mechanisms: biochemical communication and electrical communication. Biochemical communications involve messenger molecules, mostly proteins or small peptides, that target specific regulatory or metabolic networks responsible for performing and regulating specific physiological processes. The messenger molecules may lead to initiation/modification/termination of the targeted network resulting in concomitant changes of the intended physiological functions. Biomolecules used for biochemical communications and signaling pathway networks inside and between living cells are very diverse in nature, they include neuro-transmitters, glia-transmitters, proteins and peptides, cytokines, chemokines, miRNA and long non-coding RNA, calcium ions, reactive oxygen radicals and many others. Biochemical signaling between cells can be initiated by many ways. Direct contact between cells is required for many crucial processes, like cell proliferation and cell migration, during development. Immune cells communicate and share information with each other through touch, possibly via immune-effector/immune-mediator molecules transported across cell pores upon contact with each other. Neuroglia cells, also, use touch and direct contact with each other and with target organs/tissues. Cytonemes constitute another mechanism of communication between cells. They exist on surface of cells producing signaling proteins and get in direct contact with target cells that receive these signaling proteins (Figure 14).
Figure 14.Regulation of signal transduction networks
Electrical communications, on the other hand, are mediated by electrical impulses generated inside neurons within the brain or the spinal cord. Inside the brain, impulses propagate along intra and interstitial nerve tracts/bundles to initiate/regulate/coordinate neurological functions. Electrical impulses participating in regulation of physiological functions of peripheral organs, e.g., muscles and viscera, are generated in neurons located within the anterior and posterior horns of the spinal cord and propagate along peripheral nerves to their destinations. Electrical signal transduction occurs at the synapses or the site of contact between neurons.
At the synapse, the plasma membrane of the signal-generating neuron, the presynaptic neuron, comes into close apposition with the membrane of the target, signal-receptor or postsynaptic neuron. In many synapses, the presynaptic part is located on an axon, but some postsynaptic sites are located on a dendrite or soma. Astrocytes also exchange information with the synaptic neurons, responding to synaptic activity and, in turn, regulating neurotransmission. There are two different types of synapses:
1. Chemical Synapses where the electrical signal in the presynaptic neuron results, via activation of voltage-gated calcium channels, in release of neurotransmitter stored in synaptic vesicles. Binding of released neurotransmitters to signal receptors of the plasma membrane of the postsynaptic cell results in initiation of a signal response, secondary messenger pathway, that may initiate or inhibit specific functions of the target or the postsynaptic neuron. Chemical synapses are classified according to the neurotransmitter released at their presynaptic axon terminals into many types: cholinergic, adrenergic, glutamatergic and GABAergic synapses.
2. Electrical Synapses where presynaptic and postsynaptic neuronal membranes are intimately connected by gap junctions capable of transmitting nerve impulses from the presynaptic neuron to the postsynaptic neuron where release of specific neurotransmitter signal effector molecules takes place.
Ephaptic coupling, in which communication between adjacent neurons occurs via indirect electric influences, is another form of electrical communication in the nervous system distinct from synaptic communication. The exact mechanisms of ephaptic coupling is not clear, it may be mediated by mutually generated local electric fields or via direct exchange of ions or neurotransmitters between adjacent neurons. Ephaptic coupling can affect the synchronization and timing of action potential firing in neurons, and is thought to be inhibited by myelination 20. Visual phototransduction is another peculiar form of electrical signal transduction by which light photons are converted via a cascade of complex biochemical reactions, involving G-protein coupled receptors, into electrical signals in the rod cells and cone cells, which are the photoreceptor cells involved in vision, and the photosensitive ganglion cells of the retina of the eye 21.
However, irrespective of the type or nature of electrical communications between nerve cells, it must be emphasized that electrical communications, in essence, are a different form of biochemical communications characterized by being instantaneous effectors of certain physiological processes that have to be conducted immediately in response to specific stressful/adaptive physiological or environmental alterations. Nerve impulses can release neurotransmitter/signal molecules stored in synaptic vesicles located in the axon terminal of the presynaptic neurons. Conversely, they can cross the synaptic cleft separating the axon of the presynaptic neuron from the dendrites of the postsynaptic neuron. In either case, final signal transduction occurs in response to signal neurotransmitter chemical molecules released from storage vesicles in either the pre-synaptic or post-synaptic locations.
Though signal transduction pathways/networks are key regulators of nearly all cellular activities, they, in turn, work under strict control exerted by genomic regulatory networks that define the hierarchical organization and structural components of these pathways, and the framework of their functional specificities. The details of this genomic control over signaling pathway networks, like that exerted upon other network categories, are vague and incomprehensible. Protein receptors and inciting protein molecules, as proteome components mediating these pathways, are coded and synthesized by structural genes supervised by regulatory genes whose actions are controlled by master genes. Master genes are the housekeeping genes responsible, primarily, for ensuring and maintaining the three basic features of the cell genome, viz. genome identity/genome integrity/genome stability. Few master genes, e.g., those responsible for replication proofreading and DNA repair, have been characterized but the major portion of them, e.g., those responsible for organization and synchronization of signaling and developmental networks, are still indeterminate and transcendent. As mentioned before, full-scale elucidation of the detailed mechanisms of the master genomic regulatory system might be, hopefully, achieved after complete characterization of the structural organization and the functional spectrum of all components of the genetic material in living cells.
10.5.1.Characteristics of Signal Transduction Mechanisms
The peculiar specificities of cellular responses induced by different signal transduction processes are defined and regulated by a variety of molecular signaling mechanisms. Though each mechanism acts in a predefined manner to initiate/mediate/regulate the required physiological function(s), currently defined mechanisms of signal transduction have common features that characterize performance profiles of signaling networks.
1. Many signaling networks may act synergistically to achieve activation/suppression of multiple genes for cellular activities requiring participation of products of many different genes.
2. Single key signal transduction processes can induce alteration of expression profile of many different genes leading to generation/initiation of multiple related physiological outcomes.
3. Some cellular responses may be induced by multiple signaling pathways that get integrated at the signal receptor protein, generate a second messenger that forms with specific microRNA molecules specific transcription factor for genes codeing proteins mediating these responses.
4. Changes in energy kinetics of specific signal protein/signal molecule or of signal receptor protein can result in generation of distinct cellular responses. Alteration of physiological processes induced by changes in temperature/light/hydration state probably results due to kinetic changes of the signal networks regulating these processes.
5. Different cellular responses to signal transduction changes induced by the same signal protein can happen depending on the functional spectrum of the generated second messenger protein.
6. A specific signal transduction process can induce different outcomes in different tissues, in different cell types or even in certain groups of cells within a homogeneous tissue. These differential cellular responses are of utmost importance during development where groups of cells of certain regions begin to proceed along different fates, e.g., apoptosis or differentiation for instance.
7. Signaling pathway networks work in accord with general mechanisms that govern biological processes in living cells. These mechanisms include, for instance, cause and effect responses and feedback control.
8. Different cell types can use the same signaling molecules for mediation of distinct cell-specific functions. For instance, neurons use cytokines and chemokines, which are the main types of signaling molecules used by immune cells, and immune cells use many types of neurotransmitters and glia-transmitters specific for neuronal functions.
9. Some physiological processes are mediated by simultaneous integration of functions of many cell types, each performing a specific signaling role. Examples of cells that use these complex signaling processes are immune T cells and epithelial cells of the intestinal mucosa and the skin. Many neurons, for example, use multiple different neurotransmitters at the same time for performing different functions.
10. Many cell organelles, inside and outside cells, use elaborate, and have complex, signaling mechanisms. Mitochondria, for instance, have multiple signaling communications with other cell organelles and structural components, in the nucleus and in the cytosol, to regulate ATP production, apoptosis and homeostasis of calcium in the cell. Cilia are pivotal organelles indispensable for many vital physiological functions. They are primary receivers of chemical and mechanical signals necessary for hearing, smell, sight, blood flow, urine flow and cartilage pressure inside joint cavities.
11. Depending on the spatial effects of generated signal transductions, signaling pathways are classified into many different types: 1. intracrine signals produced by target cells and exert their effects within the same cell. 2. autocrine signals produced by the target cell, secreted to the extracellular space and affect the target cell itself, and sometimes close by cells, via cell surface receptors. 3. juxtacrine signals produced by signal generating cells, transmitted along cell membranes via protein or lipid components integral to the membrane, and target either the emitting cell or cells adjacent cells. 4. paracrine signals target cells in the vicinity of the emitting cell. 5. endocrine signals are generated by specific cells and target distant cells.
10.5.2.Important Signal Transduction Networks
An exceedingly large number of signal transduction networks exist in living organisms, both in intracellular components as well as in extracellular compartments in multicellular organisms [Table 2].
Table 2. Important signaling transduction networks| 1. Wnt signaling pathways |
| 2. Hedgehog signaling pathway |
| 3. The transforming growth factor beta (TGFB) pathway |
| 4. cAMP-dependent signaling pathway |
| 5. Redox signaling pathways |
| 6. Lipid signaling pathways |
| 7. Retinoic acid signaling pathway |
| 8. The JAK-STAT signaling pathway |
| 9. The MAPK/ERK pathway |
| 10. mTOR (mammalian target of rapamycin) pathway |
| 11. Hippo signaling pathway |
| 12. The Notch signaling pathway |
| 13. Calcium signaling pathways |
| 14. Apoptosis signaling pathways Extrinsic Caspases signaling pathways Intrinsic Caspases signaling pathways(Initiator/Executive Caspases signaling pathways) Fas death receptor signaling pathwaysTNF Signaling Pathways |
| 15. Cytokine and NF-κB signaling pathway |
| 16. STING-dependent signaling pathway |
| 17. Nitric oxide signaling pathway |
| 18. PI 3-Kinase/Akt Signaling pathway |
| 19. Epinephrine Signal Transduction Pathway |
| 20. Insulin signaling pathways |
| 21. Estrogen signaling pathway |
| 22. Oncogenic Signaling Pathways:The PI3K-Akt and Ras-ERK pathways VEGF mediated signaling pathway FAK-Src signaling pathwayAndrogen Receptor (AR) Signaling The Hippo-YAP signaling pathway |
10.5.2.1. Wnt Signaling Pathways
The Wnt signaling pathways are a group of signal transduction pathways that pass signals from outside of a cell, through cell surface receptors, to the inside of the cell. Three Wnt signaling pathways have been characterized: the canonical Wnt pathway, the non-canonical planar cell polarity pathway, and the non-canonical Wnt/calcium pathway. Wnt signaling pathways are activated by the binding of a Wnt-protein ligand to a specific protein receptor, which passes the generated signal to a specific signal receptor protein inside the cell. The canonical Wnt pathway leads to regulation of gene transcription, the non-canonical planar cell polarity pathway regulates the cytoskeleton that is responsible for the shape of the cell, and the non-canonical Wnt/calcium pathway regulates calcium homeostasis inside the cell. Wnt signaling pathways can work as paracrine signals targeting nearby cells or autocrine signals targeting the same cells 22. Wnt signaling play critical roles during embryonic development where they mediate, coordinate and control essential cellular/tissue/organ processes that constitute the basic framework of development. These processes include: body axis patterning, cell differentiation and specification, cell proliferation, and cell migration23. These processes are necessary for proper formation of important tissues embryonic and fetal tissues including bone, heart, and muscle. Also, enhancing cell's sensitivity to insulinvia increasing glucose uptake from the bloodstream specifically in skeletal muscles has been found to be partially mediated by activation of a specific Wnt/β-catenin signaling. Mutations affecting genes responsible for coding protein components of these pathways result in development of a variety of diseases, including breast/prostate cancer, glioblastoma, type II diabetes mellitus, congenital malformations, and many others (Figure 15).
Figure 15.Wnt signaling pathways during neurological development
10.5.2.2. Hedgehog (Hh) Signaling Pathway
The Hedgehog signaling pathway network is one of the key regulators of embryonic development where it transmits to embryonic cells regulatory commands required for proper development. Different parts of the embryo have different concentrations of hedgehog signaling proteins. Humans, and other mammals, have three distinct Hedgehog proteins, each is encoded by a different gene and expressed during specific times of development in specific embryonic/fetal cell types: Desert hedgehog (DHH), Indian hedgehog (IHH), and Sonic hedgehog (SHH). The release, diffusion and binding of these proteins to the transmembrane domain proteins is controlled by various other proteins including Skinny hedgehog (Sit), Dispatched (Disp), Tout-velu (Ttv) and Hedgehog-interacting protein (Hip). In view of their vital roles during development, these pathways are controlled by genomic regulatory networks comprising transcriptional enhancer and suppressor regulatory species of microRNAs 24. Mutations leading to deficient synthesis of protein components of the pathway result in developmental defects affecting many organs including the brain, the lungs, the gastrointestinal tract and the skeleton. The Hh pathway, probably, has an important role in regulating adult stem cells involved in maintenance and regeneration of adult tissues. Aberrant activation of the pathway in adult life has been implicated in development of many types of malignant tumors, e.g., basal cell carcinoma 25, 26.
Sonic hedgehog protein (SHH) plays key roles in regulating organogenesis, such as growth of digits on limbs and organization of the brain. It is encoded by SHH gene located on long arm of chromosome 7, is a critical morphogen that diffuses to form a concentration gradient and has different effects on the cells of the developing embryo depending on its concentration. Mutations of the SHH gene are the leading causes of holoprosencephaly and other midline cerebral defects. SHH remains important in the adult life since it controls cell division of adult stem cells and has been implicated in development of some cancers. Desert hedgehog protein (DHH) is encoded by the DHH gene located on the long arm of chromosome 12. Defects in this protein have been associated with partial gonadal dysgenesis (PGD) accompanied by mini-fascicular polyneuropathy. It may be, also, involved in gonadal differentiation in males and perineurial development. Indian hedgehog protein (IHH) is encoded by the IHH gene located on the long arm of chromosome 2. It specifically plays a major regulatory role in bone growth and differentiation and is involved in regulating the differentiation, proliferation and maturation of chondrocyte especially during stage of endochondral ossification. Its effects are regulated by feedback control of a parathyroid hormone-related peptide (PTHrP). Known mutations in this gene are linked to many developmental bone malformations including brachydactyly type A1 and acro-capito-femoral dysplasia 27 (Figure 16, Figure 17).
Figure 16.Hedgehog signalling pathway during development
Figure 17.Sonic-hedgehog signaling pathways during neurological development
10.5.2.3. The Notch Signaling Pathway
The notch signaling pathway is an important cell signaling network responsible for, and participating in, conduction of many vital physiological processes during development and in adult life of human beings as well as of other species 28. The Notch receptor is a single-pass trans-membrane receptor protein. Four types of these intracellular Notch receptors have been characterized, viz. Notch-1, Notch-2, Notch-3, and Notch-4. The Notch signal protein spans the cell membrane, with part of it inside and part outside the cell membrane. The signaling cascade consists of Notch ligands, Notch receptors, and intracellular proteins that convey the notch signal to the cell's nucleus. The Ligand binds to the extracellular domain and induces proteolytic cleavage and release of the intracellular domain of the Notch receptor which, then, forms a multi-protein complex with transcriptional co-activator proteins to initiate transcription of Notch-induced gene target genes 29.
The transcriptional co-activator proteins of the Notch pathway, e.g., members of Mastermind-like proteins encoded by the Mastermind-Like (MAML1) gene family, can modify the Notch signaling in different cell types based on their own expression levels and differential activities, thereby contributing to the diversity of the biological effects resulting from Notch activation. The functional spectrum of Notch signaling pathways is considerably wide and very diverse, it includes: cell differentiation processes during embryonic and adult life, control and synchronization of different developmental stages of the heart, regulation of development of neural functions, hematopoiesis, bone formation, functional differentiation of both the endocrine and the exocrine portions of the pancreas, differentiation of immune T-cells, control of development of the mammary glands, functional differentiation of secretory and absorptive parts of the gut, regulation of development of the vascular endothelium and angiogenesis.
In view of the wide functional spectrum of Notch signaling network in cellular activities during development as well as in adult life, mutations affecting genes encoding protein components of the pathway, or genes encoding transcriptional coactivator proteins, result in aberrant functioning and defective regulation of processes mediated by the pathway and have been implicated in pathogenesis of many disease categories including cancer, T-cell acute lymphoblastic leukemia, tetralogy of Fallot, Alagille syndrome, and many neurodegenerative conditions like multiple sclerosis and leukoencephalopathy 30.
10.5.2.4. The Transforming Growth Factor Beta (TGFB) Signaling Pathway
The transforming growth factor beta (TGFB) signaling pathway is involved in many cellular processes in both the adult organism and the developing embryo, including cell growth, cell differentiation, apoptosis, cellular homeostasis and other functions. The mechanism of action of TGFB signaling pathway, like most other signaling pathways, involves activation of the pathway via binding of a ligand protein to the protein receptor, and generation of transduction cascade followed by formation of protein complexes that act as transcriptional factors participating in the regulation of expression of target gene 31. Activation of expression of target genes is followed by initiation of synthesis of proteins necessary for performing the physiological functions controlled by the pathway. Conversely, suppression of expression of the target genes and cessation of protein synthesis occurs when the functions are accomplished and no more proteins are required (Figure 18).
10.5.2.5. Redox Signaling
Reactive oxygen species (ROS), e.g., peroxides, superoxides, hydroxyl radical, and singlet oxygen, are formed as a natural byproduct of normal metabolism of oxygen and have important roles in cell signaling and homeostasis. Environmental stresses, e.g., high temperature, overexposure to ionizing radiation and UV ray, cause marked increase in levels of ROS, oxidative stress, leading to significant oxygen-provoked damage to cell structures. ROS such as hydrogen peroxide (H2O2) are thought to initiate signaling by broadly and nonspecifically redox-modifying signaling molecules, suggesting that H2O2 signaling may be distinct from other signal transduction pathways. H2O2 signaling is under control of what appears to be a typical signal transduction cascade that connects the respiratory chain to the mitochondrial inter-membrane space-localized conserved Syk pathway and results in a focused signaling response in diverse cell types (Figure 19). This reveals a mechanism that allows the respiratory chain to communicate with the remainder of the cell in response to ROS 32.
10.5.2.6. Cyclic Nucleotide Signaling Pathways
The cyclic nucleotides, cyclic adenosine monophosphate (cAMP) and cyclic guanosine monophosphate (cGMP), have been recognized as important signaling molecules within cells. The cAMP-dependent pathway, a G protein-coupled receptor-triggered signaling cascade, is one of several cyclic nucleotide signaling pathways used, primarily, in intracellular communications. The pathway works by activating protein kinase A and, thus, further physiological processes mainly depend on cAMP-dependent protein kinase, which vary according to the type of the signal transduction generating cell. Under normal physiological conditions, cyclic nucleotides regulate a myriad of biological processes such as cell growth and adhesion, energy homeostasis, neuronal signaling, regulation of heart rate, of cortisol secretion, catabolism of glycogen and lipids, and muscle relaxation. Activation of cyclic nucleotide signaling pathways can be induced by a number of molecular mechanisms including induction of cyclic nucleotide synthesis, inhibition of cyclic nucleotide degradation, or activation of cyclic nucleotide receptors. Altered or defective regulation of these nucleotide pathways is capable of inhibiting cell proliferation and activating apoptosis, and has been observed in a number of pathophysiological conditions, including development and progression of cancer 33.
10.5.2.7. Lipid Signaling
Lipid signaling refers to biological signaling events involving lipid messengers that binds to a protein target, such as a receptor kinase or phosphatase, which in turn mediate the effects of these lipids on specific cellular responses. Common lipid signaling molecules include lysophosphatidic acid, sphingosine-1-phosphate, platelet activating factor, and arachidonoyl ethanolamine. Lipid signaling is thought to be qualitatively different from other classical signaling networks because lipids can freely diffuse through membranes. Consequently, lipid messengers cannot be stored in vesicles prior to release and are biosynthesized on demand at their intended site of action. As such, many lipid signaling molecules cannot circulate freely in solution but, rather, exist bound to special carrier proteins in serum. In view of the essential functions of lipids as integral structural components of bio-membranes and as metabolic substrates for essential hormones, the roles played by lipids as intracellular and extracellular messengers allow them to have critical regulatory roles on the fate of cells in normal physiological states as well as in disease conditions. Disturbed regulation of lipid signaling pathways has been linked to many pathological states including metabolic disorders, cardiovascular and degenerative diseases, chronic inflammatory conditions, and cancer 34.
10.5.2.8.Retinoic Acid Signaling Pathway
Retinoic acid is a metabolite of vitamin A (retinol) that mediates the functions of vitamin A required for growth and development in humans and other higher animals. During early embryonic development, retinoic acid generated in a specific region of the embryo helps determine position along the embryonic anterior/posterior axis by serving as an intercellular signaling molecule that guides development of the posterior portion of the embryo. It acts through Hox genes, which ultimately control anterior/posterior patterning in early developmental stages.The key role of retinoic acid in embryonic development mediates the high teratogenicity of retinoid pharmaceuticals used for treatment of cancer and acne. Oral megadoses of pre-formed vitamin A and retinoic acid itself also have teratogenic potential by this same mechanism 35.
Retinoic acid is an essential component of cell-cell signaling during vertebrate organogenesis. In early development, retinoic acid functions as a trunk organizer by providing an instructive signal for posterior neuroectoderm and foregut endoderm and a permissive signal for trunk mesoderm differentiation. At later stages, retinoic acid contributes to the development of the eye and other organs. Recent studies suggest that retinoic acid acts primarily in a paracrine manner and provide insight into the cell-cell signaling networks that control differentiation of pluripotent cells 36.
10.5.2.9. The JAK-STAT Signaling Pathway
The JAK-STAT signaling pathway transmits information from chemical signals outside the cell, through the cell membrane, into promoters of target genes in the nucleus, leading to activation of expression of target genes. The JAK-STAT system is a major signaling alternative to the second messenger system. It consists of three main components: a receptor, Janus kinase (JAK), and Signal Transducer and Activator of Transcription (STAT). Many JAK-STAT pathways are involved in regulation of the immune system. The receptor can be activated by a variety of signal transducing compounds including interferon, interleukin, growth factors, and other chemical messengers. This activates the kinase function of JAK, which auto-phosphorylates itself. The STAT protein then binds to the phosphorylated receptor, where STAT is phosphorylated by JAK. The phosphorylated STAT protein binds to another phosphorylated STAT protein (dimerizes) and translocates into the cell nucleus where it exerts its effects on target genes. The pathway is implicated in regulation of a number of crucial basic cellular functions including cell growth, differentiation and apoptosis 37.
The finding that Janus kinases (JAKs) and signal transducers and activators of transcription (STATs) signaling play important regulatory roles in many essential normal cellular activities, including proliferation/invasion/survival, as well as many pathological cellular disorders like inflammation and immunological abnormalities led to the use of JAK/STAT pathway inhibitors for treatment of some autoimmune diseases such as rheumatoid arthritis and psoriasis. More importantly, was the finding that mutation-induced defects in genes encoding components of this pathway are implicated in development of immune deficiency disorders and cancer. The roles of these mutations in carcinogenesis might be dependent on, or linked to, still uncovered roles of non-coding RNA transcripts in this regard. Currently, many researches are aiming at targeting the JAK/STAT pathway as a promising therapeutic strategy in treatment of some cancers, e.g., prostate cancer, hematopoietic malignancies and sarcomas 38.
10.5.2.10. The MAPK/ERK Pathway
The MAPK/ERK pathway (also known as the Ras-Raf-MEK-ERK pathway) is a signaling network composed of a chain of proteins in the cell that communicates a signal from a receptor on the cell surface to specific target genes in the cell nucleus. Like similar signaling pathways working through the same mechanism, the pathway starts when a signaling molecule binds to the receptor on the cell surface and ends when the target gene is activated and expresses specific protein/enzyme/microRNAs required for performing certain cellular functions. The pathway, composed of many proteins including MAPK (mitogen-activated protein kinases) is involved in regulation of cell division, and mutation-induced synthesis of defective components of the pathway can result in development of many cancers.
The Erk signaling pathway seems to play vital functions in cellular activities. Erk signaling is essential for self-renewal of human embryonic stem cells and plays critical roles in maintaining genomic stability. Lack of Erk, induced by pharmacological Mek inhibitors leads to rapid telomere shortening and genomic instability, in association with misregulated expression of pluripotency genes, reduced cell proliferation, G1 cell-cycle arrest, and increased apoptosis. Erk signaling is also required for the activation of differentiation genes but not for the repression of pluripotency genes during ESC differentiation 39.
10.5.2.11. mTOR Signaling Pathway
mTOR, mechanistic Target Of Rapamycin, is a serine/threonine protein kinase encoded by the MTOR gene. It is a master growth regulator involved in regulation of cell growth, cell proliferation, cell motility, cell survival, protein synthesis and autophagy. Presumed mechanism(s) of action of the pathway include sensing and integrating diverse nutritional and environmental cues, including growth factors, energy levels, cellular stress, and amino acids. It couples these signals to the promotion of cellular growth by phosphorylating substrates that potentiate anabolic processes such as mRNA translation and lipid synthesis, or limit catabolic processes such as autophagy 40. Defective functionaing of this network has been implicated in development of many human diseases such as diabetes, obesity, depression, and certain cancers. mTOR is implicated in the failure of a pruning mechanism of the excitatory synapses in autism spectrum disorders 41. Moreover, disruption of mTORC1 protein directly inhibits mitochondrial respiration, and hyperactivity of the signaling pathway has been demonstrated in brains of individuals suffering from Alzheimer’s disease42.
Altered regulation of mTOR functional/regulatory network seems to plays crucial roles in carcinogenesis 43. Over-activation of mTOR signaling significantly contributes to the initiation and development of tumors and mTOR activity was found to be deregulated in many types of cancers including breast, prostate, lung, melanoma, bladder, brain, and renal carcinomas. Reasons for constitutive activation are several. Among the most common are mutations in tumor suppressor PTEN gene. Additionally, mTOR activity is deregulated in many cancers as a result of increased activity of PI3K or Akt. Similarly, overexpression of downstream mTOR effectors 4E-BP1, S6K and eIF4E leads to poor cancer prognosis. Also, mutations in TSC protein that inhibits the activity of mTOR may lead to a condition named tuberous sclerosis complex, which exhibits as benign lesions and increases the risk of renal cell carcinoma 44. Increasing mTOR activity was shown to drive cell cycle progression and increase cell proliferation mainly due to its effect on protein synthesis. Moreover, active mTOR supports tumor growth indirectly by inhibiting autophagy. Constitutively activated mTOR functions in supplying carcinoma cells with oxygen and nutrients by increasing the translation of HIF1A, supporting angiogenesis, and up-regulating expression of glycolytic enzymes leading to increased growth rate-activation of glycolytic metabolism of malignant cells. These observations regarding the role of mTOR in promotion of carcinogenesis led to designing researches aiming at development of mTOR inhibitors as anticancer agents 45.
10.5.2.12. Hippo Signaling Pathway
The Hippo signaling pathway, also known as the Salvador/Warts/Hippo (SWH) pathway, controls organ size through the regulation of cell proliferation and apoptosis. The pathway takes its name from one of its key signaling components, the protein kinase Hippo (Hpo), a member of a family (Ste-20 family) comprising a highly conserved group of serine/threonine kinases that regulate several cellular processes, including cell proliferation, apoptosis, and various stress responses. These kinases are known regulators of cell cycle progression, growth, and development. Mutations in the Hippo gene lead to excessive overgrowth of tissues and a hippopotamus-like phenotype. The Hippo signaling pathway is involved in restraining cell proliferation and promoting apoptosis. As many cancers are marked by unchecked cell division, this signaling pathway has become increasingly significant in the study of human cancer 46.
10.5.2.13. Calcium Signaling
Calcium ions constitute crucial cellular signaling system that mediates vital physiological functions in nearly every cell in the body. This is attributed, mostly, to their allosteric regulatory effects on enzyme and protein components of metabolic networks. The functional spectrum of calcium signaling pathway encompasses a wide range of vital cellular processes including modulation of neurotransmission and plasticity of synaptic functions, muscle contraction, regulation of permeability of ion channels and synchronized functioning of ion pumps, regulation of coagulation processes, regulation of cell movement and movement of intracellular and extracellular cell organelles, regulation and control of fertilization, and regulation of cell growth and proliferation 47 (Figure 20).
10.6. Apoptosis Networks
Life and death go hand in hand all through the life span of living creatures with actual life starting with fertilization and actual death signaling the end of the life span of the organism. However, in multicellular organisms, biological death can happen on cellular level where groups of cells are selectively driven to death, where functional termination of all cellular processes and structural disintegration and decomposition of cell constituents occur. Death of groups of living cells in multicellular organisms does not happen spontaneously, rather, it is induced by a multitude of pathogenic factors that can adversely affect cellular activities. Factors that can cause cell death include external environmental pathogenic conditions like infections, irradiation and intoxication, as well as internal pathogenic conditions that adversely alter cellular homeostasis leading to intolerable damage to vital life processes in the cell. However, cell death might be intentionally induced and programmed by the genetic system that regulates life activitiea of the cell. This programmed cell death, or apoptosis, might be induced by a variety of pathogenic as well as non-pathogenic conditions for different reasons. Non-pathogenic conditions associated with, and inducing, apoptosis involve, mostly, developmental processes where groups of cells/tissues/minute structures have to be removed from, or remodeled within, developmental fields for proper organogenesis. Examples of developmental processes that involve apoptosis include: 1. elimination of transitory organs and tissues, e.g., removal of phylogenetic vestiges like pronephros and mesonephros. 2. remodeling of tissues, e.g., of limb buds and formation of digits and toes 48.
There are two main categories of pathways that initiate and mediate apoptosis: 1. extrinsic pathways where the pathway starts by extrinsic signal transduction triggered by many factors including TNF-alpha (Tumor necrosis factor-alpha), Fas ligand which forms the death-inducing signaling complex (DISC) upon ligand binding, TWEAK (TNF-related weak inducer of apoptosis), TRAIL (TNF-Related Apoptosis Induced Ligand), and TL1A (TNF-like ligand 1A). 2. intrinsic pathways initiated by many proteins of the Caspase family (Pro-Caspase 3, Caspase 6, Caspase 7, Caspase 9, etc) 49 .
10.6.1. Cascade of Signaling Mechanisms of Apoptosis
In healthy cells, phosphorylated Bad is sequestered in the cytoplasm by the 14-3-3 protein, and Bcl-2 and Bcl-xL bind to the pro-apoptotic Bax and BAK proteins to inhibit apoptosis. When cytoplasmic levels of free Bad increase, Bcl-2 and Bcl-xL bind to Bad and release Bax and BAK. Bax and BAK, or processed forms of these proteins, can then insert into the mitochondrial membrane, compromising its integrity. Loss of mitochondrial integrity results in the release of pro-apoptotic proteins including Cytochrome c, Smac/Diablo, HTRA2/Omi, Apoptosis-Inducing Factor (AIF), and Endonuclease G. In the cytoplasm, Cytochrome c interacts with APAF-1, which recruits Pro-Caspase-9 to form the apoptosome. Within this complex, Caspase-9 is processed and activated. The intrinsic pathway of caspase activation can also lead to Caspase-2 activation. Following DNA damage, p53 induces the expression of PIDD (p53-induced protein with a death domain), which associates with the CRADD/RAIDD adaptor protein and Pro-Caspase-2 to form the PIDDosome. Formation of this complex leads to the cleavage and activation of Pro-Caspase-2. Once activated, the initiator caspases, Caspase-2, Caspase-8, Caspase-9, Caspase-10, cleave downstream effector caspases, Caspase-3, Caspase-6, Caspase-7, which then promote the ordered disassembly of the cell by targeting a number of critical cellular proteins, including structural proteins, DNA repair proteins, and proteins involved in signal transduction pathways 50, 51.
10.6.2. Characteristic Features of Apoptosis
1. Apoptosis is a highly regulated and controlled vital life process that confers advantages during an organism's lifecycle. In addition to its importance as a biological phenomenon, defective apoptotic processes have been implicated in a wide variety of diseases. Excessive apoptosis causes atrophy, whereas an insufficient amount results in uncontrolled cell proliferation, such as cancer. Some factors like Fas, Caspases (C-cysteine rich, asp-aspartic acid moiety containing, ase-proteases) etc. promote apoptosis, while others like members of Bcl-2 inhibit apoptosis.
2. Apoptosis is characterized by maintenance of intact cell membranes during the suicide process so as to allow adjacent cells to engulf the dying cell so that it does not release its contents and trigger a local inflammatory reaction.
3. Signals that can trigger apoptosis can include lineage information, damage due to ionizing radiation or viral infection or extracellular signals. Extrinsic signals may either suppress or promote apoptosis, and the same signals may promote survival in one cell type and invoke the apoptosis cascade in others.
4. The intrinsic pathway of caspase activation is initiated by events such as DNA damage, growth factor withdrawal, or loss of contact with the extracellular matrix. These events ultimately lead to changes in the integrity of the mitochondrial membrane, which is regulated by Bcl-2 family proteins. The balance between pro- and anti-apoptotic Bcl-2 family members determines whether or not a cell will undergo apoptosis.
5. Apoptosis plays a crucial role in maintaining integrity and stability of the genome. Heavily mutated cells, duo to exposure to mutagenic insults, can be driven along many different destinations that devastate their functional performance. They may suffer deterioration of their physiological potential with consequent development of disease states due to failure of their life activities due to many causes like disruption of their oxidative-phosphorylation networks responsible for ATP energy production. Cells exposed to carcinogenic mutational events can undergo malignant transformation and form cancer. Also, chromosomal breaks and rearrangements can follow exposure to clastogenic agents, like many drugs and irradiation, and result in development of a wide variety of diseases including immunodeficiency and cancer. Apoptosis of heavily mutated cells acts as a prophylactic biological mechanism preventing development of devastating pathogenic states that result, ultimately, in devastation of the genome of the cell.
10.7 Oscillatory Rhythm Networks
Life activities in living cells, and living creatures, are not one-way irreversible processes. They occur recurrently and repeatedly over and over all through the life span of the organism. The dynamic nature of these activities accounts for the existence of precise mechanisms responsible for adapting the wide spectrum of biological functions to the changing conditions of their environment. Life activities depend on, and are regulated by, predictable light:dark cycles and dependent rhythms of nutrient availability. Accordingly, a vast majority of living organisms, ranging from archaea to humans, have adopted molecular mechanisms to anticipate and respond to daily metabolic rhythms. Central to this timing mechanism in mammals is a cell-autonomous molecular circadian oscillator based on transcription-translation feedback loops mediated by transcription activators, e.g., CLOCK, BMAL, and ROR, and transcription repressors, e.g., CRY, PER, and REV-ERB acting concert to generate daily rhythms in their own protein levels as well as thousands of target genes. For example, metabolic adaptations in response to availability or deficiency of nutrient supply represent one prominent example of these adaptations thought to temporally coordinate metabolism to the appropriate time of the day to sustain metabolic homeostasis. These adaptations are affected by many factors including feeding-fasting rhythms mediated by cell-autonomous circadian clock and feeding-fasting driven molecular programs 52.
Physiological processes occur in order to meet and fulfill the biological needs of the organism and represent spontaneous functional responses to external and/or internal demanding conditions. Most physiological processes, however, have to be properly coordinated with daily cyclic alterations of external environmental effectors like light, darkness, and temperature, where precise synchronization between internal milieu and external environment is necessary to keep proper homeostasis of life activities of living organisms. This round the clock synchronization of the rhythm of vital life processes with that of natural daily cyclic variations is mediated/regulated/maintained by a peculiar control system composed of specific set of regulatory networks referred to as circadian rhythm networks. Many other physiological processes that have no apparent cause and effect relationship with daily external environmental effectors, however, have to be properly synchronized with the continuous everlasting cyclic alterations of internal cellular conditions. These processes, also, occur in a spontaneous oscillatory rhythmic pattern, and are mediated/synchronized/maintained by a specific set of regulatory networks that might be tentatively referred to as intrinsic rhythm networks.
10.7.1 Circadian Rhythm Networks
The master clock of the circadian biological system is located in the suprachiasmatic nucleus (SCN) of the hypothalamus, and has peripherally located clocks found in most tissues. This system has an endogenous rhythmicity of approximately 24 hours. Peripheral systems are synchronized by the SCN. Melatonin hormone represents the key linkage-molecule between the SCN and the peripheral biological clocks and it is produced and secreted in a circadian fashion, its secretion being stimulated by darkness and inhibited by light 53. The core components of these regulatory network comprise a large number of genes known as circadian clock and clock-related genes. The protein products of these genes constitute the structural components of the networks necessary for the generation and regulation of circadian rhythms. The circadian rhythm oscillations of physiological processes, including behavior, reflect the cyclic functioning of the autoregulatory loops that control transcriptional-translational activities of clock and clock-related genes (Figure 21, Figure 22).
Figure 21.Human circadian rhythms
Figure 22.Genetic regulation of circadian rhythm networks
The circadian system is responsible for regulating a wide variety of physiological and behavioral activities including rest-wake cycle, cardiovascular activity, hormone secretion, body temperature and metabolism. Mutations of clock and clock-related genes have been implicated in pathogenesis of many human disorders including sleep disorders, mood disorders, and breast/endometrial/pancreatic cancer. Circadian regulation in peripheral tissues is important to maintain normal cellular functions, and a disruption of core clock genes can be damaging to the organism’s overall well-being 54.
10.7.2 Intrinsic Rhythm Networks
Intrinsic rhythm networks are responsible for regulating life activities and physiological processes initiated by rhythmic stimuli generated, on regular bases, by different components and diverse biological situations within living cells. Contrary to circadian rhythm stimuli, generated in response to daily changes of external environmental effectors, intrinsic rhythm stimuli are not triggered by external factors, rather, they are initiated by intrinsic rhythmic variations in cellular activities. The most apparent examples within this context are the synchronized neural oscillations which represent a fundamental mechanism for enabling coordinated activity during normal brain functioning and a variety of cognitive and perceptual functions 55. Neuronal networks, a subcategory of regulatory continual unceasing networks, are highly organized complex systemsresponsible for regulating cellular mechanisms underlying neuronal synchronization and electrical oscillations in the nervous system. Through controlling generated rhythmic electrical neuronal activities, they participate in processing information required for higher brain functions including behavior, cognition, and memory. Alteration of rhythmic electrical neuronal activities by various disorders of the nervous system, or disruption of structural organization of regulatory neuronal networks, caused by mutations affecting their protein components, underlies development of many pathophysiological mechanisms responsible for a wide spectrum of neurological disorders secondary to affection of learning capabilities, cognitive performance and memory processes (Figure 23).
Figure 23.Neural control of central and peripheral circadian rhythms
The exact nature of the mechanism(s) underlying rhythmic physiological activities are vaguely delineated and poorly understood. One possible cause of this situation might be the current traditional definition of the term rhythmic, since it restricts its use to rhythmic activities of neural cells and sino-atrial node rhythm, both of which are, still, enigmatic rhythmic life activities. Extending the definition of rhythmic activities to include any regularly occurring physiological processes initiated by other regularly occurring processes would add and reveal a wide spectrum of myriads of diverse physiological situations that occur in rhythmic pattern. In fact, meticulous analysis of life activities in living cells reveals that all normal physiological functions that occur on regular basis, in absence of external harmful effects, are mediated according to peculiar rhythmic patterns. Starting from scratch, gene function, beginning with sensing the need for the gene product and ending with synthesis of sufficient amounts of the normal gene product, happens in a rhythmic pattern regulated by many feedback loops to ensure initiation of transcription and continuation of translation until the required amounts of the gene product are synthesized. The rhythmic pattern of gene expression might happen as around-the-clock activity for, e.g., master housekeeping genes, or according to compelling cellular demands like metabolic and signaling transduction processes.
The same principles governing the rhythmic behavior of gene function, probably, apply to other, apparently, unrelated processes that occur all through the life span of the cell, like shortening of the telomeres with every cycle of cell division, waste disposal by lysosomes when a threshold of harmful accumulation of waste products within the cell is reached, periodic or rhythmic activation and termination of signaling functions according to physiological needs and, most significantly, apoptosis. However, whereas the rhythmic pattern of some physiological functions, e.g., wakefulness-sleeping pattern and feeding behavior, can be interpreted in view of the changing environmental conditions, e.g., daily changes of light-darkness and availability of food respectively, the majority of other recurrently occurring biological processes in the cell have no satisfactory explanation yet. Presence of sensory networks responsible for perception of deviations of intrinsic biological environment from optimal standards dictated by the classic dogma of life, and corresponding existence of executive networks responsible for initiating appropriate adaptive responses to these deviations seems reasonable theoretical hypotheses to explain the nature of rhythmic behavior of life activities, but they remain assumptive, as none of these postulated perceptive or executive networks have been defined yet.
A considerable obstacle to formulate a reasonable hypothesis capable of offering logical interpretation of the nature of the control system of rhythmic life phenomena, prior to research experimentation, stems from, and resides in, the peculiar nature of the basic structural design of biological networks. Contrary to artificially engineered networks that have a limited degree of flexibility in response to environmentally induced stresses, biological networks have an amazing degree of resilience and elasticity to changes in external environmental conditions as well as to intrinsic stressful situations. For instance, without electricity, engineered networks can not start working whereas humans are capable of withstanding thirst for days and can tolerate starvation for weeks, and some living creatures can withstand these drastic conditions for months, and even for much longer times. Another prominent example is the wide range of physiological options the living cell has in response to environmental stresses. Whereas the functional design of artificial networks is based upon, and ruled by, strict principles headed by programmed automation, biological networks are designed so as to permit a considerable degree of self automation in addition to programmed automation. Self automation allows for perception of internal and external environmental changes. It allows, also, for selecting and preferring the best physiological responses available to adapt to these changes. Furthermore, it permits repair and corrective pathways to interfere for ensuring the appropriateness of these responses.
The concept of self automation of biological networks might present a partly accepted hypothesis to interpret some of the incomprehensible aspects of the rhythmic pattern of life activities, but it is far from being satisfactory. One assumption of the hypothesis imparts to some biomolecules, essentially proteins and some microRNA classes, the ability to sense and perceive what changes are happening in the surrounding micro environment based on biochemical, biophysical, or electrical changes in the cell. Another assumption postulates the ability of these components to respond by choosing the best appropriate adaptive or corrective responses, either to maximize the benefits or to minimize the damage induced by these changes, via modulating the functions of networks responsible for mediating these responses. A third assumption of the hypothesis presumes the presence of a global discerning system, whether in the cell in unicellular organisms or on the whole organism level in multicellular organisms, capable of keeping the wide spectrum of the different aspects of life activities, including rhythmicity and periodicity, within the optimal framework defined by the classic dogma of life. Final and proper interpretation of the nature of the master control system and the underlying mechanisms that regulate life activities in living creatures, still, seems very far from being attained in view of the paucity of currently available information regarding the basic design of the genetic material of living organisms.
10.8. Executive Networks
There is no satisfactory definition or delineation of the meaning of executive networks within the global context of life activities in living cells. In essence, all types of biological networks and pathways are executive in nature in view of their roles in performing or executing specific physiological functions. Examples include the aforementioned genome preserving networks, transcription and translation regulatory networks, developmental networks, and apoptosis networks. The same concept applies, also, to all tributary and subsidiary networks and pathways responsible for executing partial reactions and mediating sequential responses within the wider functional framework of the mother network. The significance of including executive networks as a distinctive category within the classification scheme of biological networks stems from the need for, and reflects the benefits of, differentiating a considerable large number of important networks in the cell, that perform and execute essential and crucial non-metabolic functions, from metabolic networks and pathways that, probably, constitute the major proportion of biological networks in living cells.
Non-metabolic executive networks in the cell comprise a large number of functionally distinctive network groups responsible for regulating vital and critical cellular functions that do not conform to the traditional definition of metabolic processes. These diverse network groups include, for example:
1. Energy Generating Networks or the oxidative phosphorylation and glycolysis networks responsible for production of the energy source of all life activities in the cell in the form of Adenosine triphosphate or ATP molecules.
2. Signaling Networks involved in regulating important functions and critical aspects of most types of multistage networks either to ensure proper completion of ultimate network functions or to synchronize performance of different networks with shared or related functions.
3. Transport Networks responsible for regulating the transport of biomolecules to the inside and out from the cell across cell membranes through cell channels as well as transport of biomolecules, metabolites, metal and non-metal ions across membranes of cellular compartments.
4. Constructive Networks responsible for the construction and configuration of cellular compartments and cell organelles like the cytoskeleton, the cell membranes, the mitochondria, the endoplasmic reticulum, the peroxisomes and cell channels.
5. Neurogenic Networks responsible for executing, regulating and modulating the vast spectrum of varied physiological and psychological functions governed by the brain and the nervous system. The spectrum of these functions begins, at one end, with the most simple neurological reactions of simple organisms like stimulus-response reflexes, and ends with the most complicated of these functions like cognition, reasoning, learning, memory, reading and writing.
10.9. Metabolic Networks
Metabolic networks, probably, constitute the major proportion of biological networks in living cells (Figure 24). Although traditional definition of metabolism makes clear-cut distinction between anabolism, which is a targeted non-spontaneous process, and catabolism, which occurs in a more or less autonomous and spontaneous manner in normal conditions, a more plausible definition of metabolism would include any, and all, life processes within living cells. Many logical reasons for this suggestion do exist:
First, physiological processes in living organisms, irrespective of their location whether being intra cellular or extra cellular, do comprise one or, more often, both phases of metabolism. Synthesis of proteins, for instance, comprises anabolic processes, e.g., synthesis/polyadenylation of mRNA and elongation of the synthesized polypeptide chain, as well as concomitant catabolic processes, like selective removal of non-coding introns from final mRNA and degradation and recycling of mRNA after translation is over.
Second, anabolism and catabolism occur simultaneously during the course of all life processes to keep the sequences of these processes confined to, and conforming with, basic principles of thermodynamics which govern the reaction dynamics of biomolecules in living cells. Regular anabolism or synthesis of macromolecules in living cells involves use of smaller building molecules partly derived from regular catabolism or degradation of these macromolecules after performing their functions, and continuous supplies of energy bearing ATP molecules by the oxidative-phosphorylation energy generating networks are balanced by dissipation of their energy for mediation of life activities in the cell.
Third, though other network categories, that are functionally distinct from metabolic networks, like genomic regulatory networks and signal transduction networks, are presumed to have their own peculiar characteristic features, they, in essence, share metabolic networks their main feature represented by cyclic turnover of their constituents after performing their regulatory or signaling roles, respectively.
Traditional classification of metabolic networks comprises three main functional categories: anabolic networks responsible for mediating anabolism or synthesis of complex biomolecules from simpler molecules or primitive substrates, converting networks that catalytically convert certain biomolecules to other biochemically-related, reactive, derived or ready to use biomolecules within a specific reaction cycle like phosphorylation, methylation, glycosylation, oxidation reactions, and catabolic networks responsible for catabolism or degradation of large molecules to smaller molecules and disposal of biological waste products and damaged cellular components accumulating within the cell.
Figure 24.General outline of mechanism of function of metabolic networks
Anabolism, or constructive metabolic processes for synthesis of complex biomolecules or compounds from simpler molecules, involves three stages:
1. Production of simple precursors such as amino acids, monosaccharides, fatty acids and nucleotides.
2. Activation of these precursors into reactive forms using energy from ATP.
3. Assembly of these precursors into complex molecules such as proteins, polysaccharides, lipids and nucleic acids. The extent and complexity of anabolic networks differ widely as living organisms differ in their ability to construct their needs of biomolecules required for their vital processes. In this respect, organisms are classified into autotrophs, such as plants, that can construct their needs of complex organic molecules, e.g., polysaccharides and proteins, from simple molecules like carbon dioxide and water, and heterotrophs which require a source of more complex substances, such as monosaccharides and amino acids, to produce these complex molecules. Organisms can be further classified according to the source of energy needed for anabolism into photo-autotrophs that obtain their energy from sun light and chemo-autotrophs/chemo-heterotrophs that obtain their energy from inorganic oxidation reactions.
Similarly, the extent and complexity of catabolic networks differ greatly depending on the complexity of life processes of living organisms, including anabolic activities. Cells use simpler molecules released from catabolism of complex precursors to construct new precursors. They also degrade these simpler molecules further to final waste products, carbon dioxide, ammonia, urea, and acetic acid. The production of these wastes involves oxidation processes that lead to release of chemical energy partly lost as heat. The rest is used for synthesis of the energy bearing molecules adenosine triphosphate (ATP). Catabolism in living cells performs three major functions: supply of simple molecules for synthesis of more complex molecules needed for mediating cellular functions and building cell architecture, provision of energy necessary for the maintenance and growth of cells, and disposal of waste products of cellular activities. In addition, though many metabolites act as intermediate substrates, many others play important regulatory roles either within feedback loops or as constituents of signaling pathways involved in control of transcription and translation of proteins and enzymes of metabolic networks.
10.9.1 Functional Regulation of Metabolic Networks
Exquisite regulatory mechanisms of metabolic networks exist in living cells to insure proper synchronization within each network and between related networks involved in specific physiological activities. Attainment of optimal network performance represents a major target of these regulatory mechanisms. Optimal network performance includes many aspects headed by sufficient supply of energy and substrates necessary for mediating cellular functions. Disposal of outputs of metabolic pathways and appropriate communications between related networks represent other important aims of these regulatory mechanisms (Figure 25).
Figure 25.Overview of regulatory interactions involved in regulation of metabolic regulatory networks
Metabolic reactions are chemical reactions mediated by enzymes and proteins. Regulation of metabolism, and functions of metabolic networks regulating metabolic activities in living cells, is exerted primarily by changes in functions of genes responsible for synthesis of enzymes and other proteins constituting metabolic networks in the cell. Metabolic genes, or genes encoding metabolic proteins/enzymes, do not function in a persistent manner. They begin transcription when need for their encoded products arises, e.g., hyperglycemia leads through many signal transduction mechanisms to activation of insulin gene and synthesis of required insulin. Once euglycemic state is attained and no more need for insulin exists, the insulin gene is suppressed and synthesis of more insulin stops. One possible mechanism of regulation of metabolic genes has been postulated in prokaryotes after the identification of several different types of regulatory motif acting in a feedback loop pattern. The postulation is based on the assumption that metabolic genes are often regulated via a network motif known as a single-input motif in which a single transcription factor binds to the promoters of a set of genes. This allows coordinated control of the expression of entire metabolic pathways. Moreover, differing sensitivity thresholds of different promoters to the same transcription factor could allow an ordered temporal wave of expression through the pathway 56.
Metabolic reactions are organized into specific consecutive metabolic pathways in which one compound is transformed through a series of incessant steps into another compound, few simple compounds are combined to form a more complex compound, or a complex compound is degraded to its simpler constituents or to different output molecules. Metabolic pathways are conducted and executed by sequences of enzymes, each having a particular role in performing a specific structural change of the compounds involved in the metabolic pathway. Enzymes constitute the corner stone of metabolism and of life activities of all living organisms. The framework of metabolic reactions, anabolism for instance, is designed so that anabolic enzymes in the pathways perform reactions requiring energy by coupling them to catabolic and/or spontaneous reactions that release energy. In addition, the catalytic nature of enzyme dynamics allow biochemical reactions along metabolic pathways to proceed more rapidly, sometimes many fold increase in their default rates, to compensate for deficient availability or to increased requirements of particular compounds. Regulating the function(s) of metabolic networks happens either via quantitative alterations in concentrations or amounts of different components of the network, e.g., enzymes/proteins/signaling molecules/metabolic substrates, or via qualitative alterations of functional performance or reaction dynamics of the basic network components.
10.9.1 A. Quantitative Regulation of Functions of Metabolic Networks
This mode of regulation is achieved, primarily, through control of amounts of the network components participating in the specific reactions mediated by the network. Different mechanisms of quantitative regulation exist according to the nature of the network component.
1. Quantitative changes of amounts of the enzyme or protein, and of protein signaling molecules,can be achieved through different genetic regulatory mechanisms including:
a. Changes in transcription where extracellular signal molecules, e.g., hormones, proteins, neurotransmitters, can lead to signal transduction responses with ultimate activation or suppression of transcription of the gene coding for the enzyme.
b. Changes in translation output where the number of translation cycles of mRNA can be adjusted to translate enough amounts of the enzyme in normal conditions or can be increased to synthesize more amounts of the enzyme during periods of metabolic stress requiring more enzyme activity. This mechanism depends on small interfering microRNA molecules that bind to the mRNA thus stopping translation of more protein. Changes in stability of mRNA and its rate of degradation is another mechanism used in control of translation.
2. Quantitative changes of amounts of the substrate(s) or of signaling molecules participating in the reactions mediated by the network occur via many different mechanisms controlled by master or mainframe regulatory networks that synchronize related functions of different sub-networks responsible for performing specific physiological functions. For example, metabolic end products or intermediate products or metabolites of some networks are used as substrates of other networks. Similarly, signaling molecules of many pathways act, concomitantly, in regulating specific functions of other networks including metabolic networks. This pattern of metabolic regulation characterized by strict harmony in functions of different networks ensures optimal energy consumption with minimal dissipation or loss of energy resources of the cell. Additionally, it also ensures optimal consumption of all usable resources of metabolism within the cell in a near perfect cost-effective design of metabolic regulation.
10.9.1 B. Qualitative regulation of Functions of Metabolic Networks
This mode of regulationrefers to changing network performance by mechanisms that do not involve increasing or decreasing amounts or concentrations of components of the network. These mechanisms act, basically, through enhancing or decreasing activities of the enzymes mediating network reactions by various ways including:
1. Changes in structural conformations of functional domains of the enzyme resulting in more, or less, efficient binding of the substrates/reactants to the catalytic site(s) of the enzyme thus increasing or decreasing enzyme activity, respectively.
2. Changes in structural conformations of the enzyme resulting in decreased binding of competitive/inhibitory molecules to the active functional domains with consequent increase in activity, or conversely, resulting in exposure of these domains to competitive molecules with consequent inhibition of enzyme activity.
3. Changes in enzyme dynamics due to structural conformational changes induced by allostery, or allosteric regulation. Allostery refers to binding of a regulatory or effector molecule to domains of the enzyme/protein other than its active or orthosteric sites or domains. These regulatory domains are termed allosteric sites and are capable, upon binding to allosteric activator molecules, of enhancing enzyme dynamics, a phenomenon known as positive allosterism. Conversely, upon binding to allosteric inhibitor molecules, they can result in reduction in enzyme dynamics and decrease of enzyme activity, or negative allosterism. Allosteric regulation is an important regulatory mechanism of both metabolic and other network categories that comprise enzymes as part of their constituents, as it allows fine adjustment of activities of different cellular networks according to physiological requirements of the cell and the whole organism.
4. Changes in the temperature and pH of the environment of the reaction, where each of these variables is capable of affecting enzyme activity in a positive or negative pattern.
5. Changes in quaternary structure of the enzyme by covalent modifications involving covalent binding of the enzyme to other active chemical groups like hydroxylation, methylation, phosphorylation and sulfation. Covalent modifications, similar to post-translation modifications, can considerably affect enzyme activity with consequent conspicuous alterations in reaction dynamics.
6. Product inhibition, or inhibition of the enzyme by products of the reaction(s) catalyzed by the enzyme, is an important self-regulatory mechanism of most metabolic networks. It represents an efficient feedback loop adjusting and synchronizing network function with physiological needs, thus ensuring optimal substrate and energy consumption by various networks within the cell.
Functional regulation of metabolic networks might be intrinsic regulation exerted by the specific dynamics of the network, or extrinsic regulation induced by external effector molecules or reaction conditions. Intrinsic regulation involves self-regulation of the metabolic pathway(s) of the network in response to quantitative changes in levels of substrates or products of the pathway. A decrease in the amount of given product of a particular reaction, for example, can lead to increased or enhanced flux of the substrate through the pathway to compensate for the deficiency of the product. Intrinsic regulation, usually, involves positive allosterism of activities of enzymes mediating the pathway. Extrinsic regulation, on the other hand, implies regulation of particular intracellular network/pathway by external effectors which might be secreted or produced within the cell. It, also, implies regulation of intracellular networks/pathways by extracellular effectors secreted or produced by other cells in multicellular organisms. This mechanism of extrinsic regulation of metabolic networks, as well as of many other types of biological networks, constitutes a basic regulatory mechanism responsible for mediating biochemically-dependent intercellular/inter-tissues/inter-organs communications in multicellular organisms. The biochemical effectors involved in this kind of extrinsic regulation constitute a variety of biomolecules including hormones responsible for endocrinal regulation of tissues and organs, signal transducers responsible for initiation and propagation or transduction of regulatory primary and secondary signals/messengers of a wide variety of networks inside the cell including signaling pathways of genomic/transcriptomic/proteomic networks, and neurotransmitters which exert regulatory functions over all brain cells by different mechanisms including signal transduction and local neuronal hormonal control.
Xenobiotic alterations refer to a specific condition of induced changes in network dynamics and performance caused by xenobiotics. Xenobiotics comprise a large group of compounds of varying chemical composition, drugs/poisons/non-poisonous organic compounds etc, present in the environment. Xenobiotics can not be used as food or metabolic substrates by the cell, rather their accumulation within the cell(s) can harm or even kill the organism. With the exception of safe and appropriate use of synthetic drugs, exposure to xenobiotics can lead to detrimental consequences on exposed cells or organisms, the effects being dependent on the types and functional importance of involved networks, as well as on the site/nature/magnitude/effects of pathophysiological alterations induced by the effector xenobiotic compound.
10.9.2. Metabolic Disorders
The metabolic system, sometimes referred to as the metabolome, of a particular organism determines the different groups of compounds that can be dealt with by its inherent specific spectrum of metabolic networks. For any particular organism, ingested compounds might be nutrient foods, poisonous or toxic compounds, or non-noxious compounds. This distinction is species-specific characteristic of each organism depending on the structural components and the functional capabilities of its metabolome, as some nutrient compounds used by some species are poisonous for other species. Genetic defects affecting the integrity of the metabolic system of the organism can lead to deleterious consequences in one or more of the metabolic pathways that affect its biological fitness. Inborn errors of metabolism in humans are an example of the results of such genetic defects adversely altering the metabolome of humans.
Metabolic errors refer to diseases resulting from disturbed functions of metabolic networks. Both quantitative defects due to reduction in amounts of protein and enzyme components constituting the framework of the networks, as well as qualitative defects resulting from defective synthesis of these components can lead to development of functional disorders of the network with consequent pathogenesis of metabolic disorders (Figure 26).
Figure 26.Molecular defects in construction of metabolic networks and pathogenesis of metabolic disorders
Metabolic disorders are classified, broadly, into inborn errors of metabolism or congenital metabolic disorders and acquired metabolic disorder.
10.9.2. A. Congenital Metabolic Disorders
Congenital metabolic disorders or inborn errors of metabolism are genetically-determined inherited diseases caused by deleterious mutations affecting genes responsible for synthesis of proteins and enzymes constituting, and participating in, metabolic networks. These mutations result in either reduction in amounts of translated/synthesized proteins and enzymes or translation/synthesis of structurally defective proteins/enzymes. Genetic mutations that cause defective post-translation modification or defective post-translation trafficking and localization of synthesized proteins/enzymes result in functional deficiency and can have the same quantitative and/or qualitative pathogenetic consequences. In all of these conditions, disturbances in function(s) of relevant networks occur leading to pathophysiological alteration and pathogenesis of disease.
The vast majority of these errors are single gene disorders, and are inherited in an autosomal recessive pattern. Some of them, however, are inherited in a X-linked recessive mode, or much more rarely in a X-linked dominant pattern. Still, a minority of these errors are inherited in an autosomal dominant pattern, including, for example, acute intermittent porphyria, hereditary coproporphyria, most cases of erythropoietic protoporphyria and variegate porphyria. Individually, congenital metabolic errors are relatively rare genetic defects, but collectively they constitute a large class of common genetic diseases. Clinical presentation of most congenital metabolic defects makes its appearance usually during the neonatal period, during infancy or during childhood, but can occur at any time, even in adulthood depending on the pathophysiological consequences of the defect and the physiological importance of the organs or tissues whose functions are disturbed by the pathogenetic mechanism(s) of the disorder.
10.9.2. B. Acquired Metabolic Disorders
Acquired metabolic disorders include non-genetically determined metabolic disturbances that are caused by a variety of factors and pathological mechanisms. These causes include a long list of drugs that can induce deleterious alterations of many metabolic networks as a consequence of their side effects, a similar list of infections capable of altering body metabolism via different mechanisms including secretion of toxins or metabolic blocker and competitive molecules/damage to vital organs leading to deficient synthesis of protein and enzyme components of metabolic networks and pathophysiological alterations of water and electrolyte metabolism, many environmental toxins and over exposure to irradiations. Irradiations induce pathogenesis of nearly all known human disease categories, including metabolic disorders, due to their clastogenic, or chromosome breaking, effects and DNA damaging sequelae. Post-irradiation metabolic disturbances result from induced mutations affecting metabolic genes that encode protein and enzyme components of metabolic networks. Functional deterioration leading to progressive multiple organ failure ensues and accounts for the wide spectrum of post-irradiation induced diseases including cancer, immunodeficiency, neurodegeneration, suppressed hematopoiesis, organ/tissue damage presenting as radiation-induced lung disease, radiation-induced enteropathy, radiation-induced heart disease, radiation-induced cataract, and many others.
10.9.3 Pathogenesis of Metabolic Disorders
Many different pathogenetic mechanisms are implicated in initiation and development of metabolic disorders including:
1. Deficient generation, release, and transfer of energy, ATP, required for carrying out cellular activities, e.g., mitochondrial oxidative-phosphorylation defects.
2. Defective synthesis of required vital essential complex biomolecules from simple molecules, e.g., purine/pyrimidine synthesis defects.
3. Deficiency of final/intermediary product(s) of the network with resulting disturbance of physiological processes dependent on these products and development of disease, e.g., hormone deficiency, coagulation defects, and biomolecule deficiency disorders.
4. Accumulation of primary substrates or intermediary metabolites to above threshold toxic/damaging levels leading to disturbances in functions of target organs, development of pathophysiological alterations and pathogenesis of diseases, e.g., phenylketonuria, porphyrias and galactosemia.
5. Defective catabolism and breakdown of metabolic waste products. Pathological intracellular accumulation of these wastes leads to gradual and progressive failure of cellular functions and development of disease, e.g., lysosomal storage disorders.
6. Defective metabolism of essential biomolecules leading to their accumulation and excessive faulty storage within cells and tissues. Over time, progressive failure of physiological functions of affected tissues result in pathogenesis of disease, e.g., lipidoses and glycogen storage diseases.
10.9.4 Classification of Metabolic Disorders
Metabolic disorders are broadly classified into two major disease categories: genetically-determined inherited or congenital or inborn errors of metabolism and acquired non genetically-determined metabolic disturbances. Other classification schemes include classifications based on the nature of the affected/deficient/defective substrate or metabolite (Table 3), on the nature of affected tissues or organs, and on the nature of disturbed physiological functions.
Table 3. Major classes of metabolic diseases| Metabolic disease category | Examples of metabolic diseases | |
|---|---|---|
| 1 | Disorders of carbohydrate metabolism | glycogen storage diseases, diabetes mellitus, galactosemia |
| 2 | Disorders of amino acid metabolism | phenylketonuria, tyrosinemia, maple syrup urine disease, urea cycle defects |
| 3 | Disorders of organic acid metabolism | glutaric aciduria, propionic aciduria, methylmalonic aciduria |
| 4 | Mitochondrial fatty-acid oxidation defects | medium-chain-acyl-CoA dehydrogenase deficiency (MCAD), carnitine transporter deficiency |
| 5 | Lysosomal storage disorders | Gaucher's disease, Niemann Pick disease, mucolipidoses, mucopolysaccharidoses, sialidosis, |
| 6 | Peroxisomal disorders | Zellweger syndrome, infantile Refsum disease, neonatal adrenoleukodystrophy |
| 7 | Disorders of steroid metabolism | congenital adrenal hyperplasia, 5-alpha-reductase deficiency, Aromatase excess, Aromatase deficiency |
| 8 | Disorders of porphyrin metabolism | porphyria cutanea tarda, acute intermittent porphyria |
| 9 | Disorders of purine/pyrimidine metabolism | Lesch-Nyhan syndrome, hypouricaemia, orotic aciduria |
| 10 | Disorders of mitochondrial oxidative-phosphorylation | Leber’s hereditary optic neuropathy, Kearns-Sayre syndrome, Leigh disease/retinitis pigmentosa, neurogenic myopathy |
| 11 | Metabolic disorders of trace elements | Menkes disease, acrodermatitis enteropathica, X-linked cutis laxa, Wilson disease, Selenium deficiency, Selenium excess |
| 12 | Disorders of lipid metabolism | Hypercholesterolemia, hypertriglyceridemia, sphingolipidoses |
| 13 | Disorders of protein metabolism | Coagulation defects, immunodeficiency disorders |
| 14 | Disorders of vitamin metabolism | vitamin deficiency-induced disorders, vitamin excess intoxication disorders |
| 15 | Disorders of DNA repair | DNA repair defects: Fanconi anemia, ataxia telangiectasia, Bloom syndrome, xeroderma pigmentosum |
| 16 | Disorders of DNA replication | Replication protein A (RPA) deficiency Gerald et al. 2004 |
10.10. Preserving and Repair Networks of the Genome, the Transcriptome and the Proteome
Life of humans and other kinds of living organisms develops and persists in a hostile environment full of existing and continuously generated mutagens and life endangering stresses. Mutational events induced by external factors, viz. physical, chemical and biological mutagens, have widespread detrimental effects on the identity, stability and integrity of the genome as well as on the same aspects of both the transcriptome and the proteome. In addition, considerable round-the-clock damage of the components and, hence, the structural organization and functional capabilities of these basic constituents of life regularly occurs on continuous and progressive basis due to the continuously generated burden of internal mutagens produced from the diverse metabolic activities of the exceedingly large number of biochemical reactions and physiological activities of the cell. Under abnormal conditions, e.g., during infection or intoxication, marked increase in the burden of these harmful stresses is met with. These external and internal damaging situations must be dealt with promptly and efficiently before irreversible damage to basic life constituents occurs. In fact, without the existence of the amazingly powerful and efficient repair system responsible for preserving the genome-transcriptome-proteome constituents of living creatures, life would have been impossible.
Optimal performance of life activities within the cell necessitates precise cooperation and proper synchronization of the diverse functions of all components of the three basic constituents of life, viz. the genome, the transcriptome and the proteome, in the cell. Accordingly, preserving structural integrity of these constituents and their components represents a vital priority for the cell. The anti-mutation protective and repair systems of living cells comprise a wide spectrum of networks and pathways responsible, primarily, for protecting the structural integrity, and consequently the functional capabilities, of these constituents against the deleterious effects of external and internal mutational and non-mutational harmful events. They are responsible, also, for repair of resulting structural damage or alterations of their components to regain their functions. Each of the three basic life constituents in the cell has its own specific anti-mutation and/or repair network system.
10.10.1. Anti-mutation Networks of the Genome
The human genome is endowed with a spectacular multifaceted strong anti-mutation system responsible for maintaining the stability and the integrity of its components, and preserving its identity. It acts by protecting the genome from the detrimental effects of mutation and by repairing mutation-induced damage. These actions are performed by many types of networks and pathways each being responsible for mediating a damage-specific repair process via particular repair mechanisms. Genome-protection and repair system consists of many diverse groups of networks and subsidiary executive pathways performing wide spectrum of functions mediated by diverse types of effector molecules. Effector components of these networks include many types of proteins, enzymes, microRNAs and other classes of organic and inorganic components like biochemical buffers, modulators of charge transport, methylating and demethylating components, deactivating biomolecules, and antioxidant enzymes. The functional spectrum of this genome protection and repair network system encompasses vital and crucial functions comprising proofreading during DNA replication, repair of mutation-induced damage of DNA via different mechanisms including base excision repair/nucleotide excision repair/direct reversal repair/mismatch repair and recombination repair, preservation of DNA-associated protein assembly via antioxidant protective enzymes and nucleoprotein-microRNA complexes responsible for repair of oxidant-stress induced damage, prompt correction of DNA helix distortion, regulation of the cell cycle to ensure proper progression of all its dynamic stages and its completion until division to daughter cells with identical genomes, and proper sets of chromosomes, occurs (Table 4).
Table 4. Anti-mutation protective/regulatory/repair networks and pathways| Anti-mutation mechanisms of the human genome & human proteome | |
|---|---|
| Mechanism | Types & pathways & comments |
| 1. Structural organization of the genome | 1. Nuclear genome |
| 2. Mitochondrial genome | |
| 2. Structural features of DNA | Complementary strand stores genetic information |
| 3. Degeneracy of the genetic code | Multiple point mutations might occur without affecting synthesized protein |
| 4. Nuclear localization of DNA | Physical protection of nuclear genome |
| 5. DNA-associated proteins | 1. Physical barriers |
| 2. Biochemical buffers | |
| 3. Deactivating biomolecules | |
| 4. Modulation of charge transport | |
| 5. Limitation of DNA helix distortion | |
| 6. Replication proofreading system | Prophylactic pathway during DNA replication |
| 7. Genetic repair systems | A. Nuclear DNA repair |
| 1. Base excision repair | |
| 2. Nucleotide excision repair | |
| 3. Direct reversal repair | |
| 4. Mismatch repair | |
| 5. Recombination repair | |
| B. RNA repair/editing system | |
| Post-transcription repair/editing of some mRNA defects via guide RNA (gRNA)/editosome complex | |
| C. Mitochondrial DNA (mtDNA) repair | |
| 8. Protein repair systems | Correction of post-translation protein misfolding/aggregation by chaperones |
| 9. Silencing of transposons by piwiRNA | Reduces transposon-induced mutations duringdevelopment |
| 10. Antioxidant enzyme systems | |
| 11. Apoptosis | Prophylactic pathway against spread of mutations of heavily or lethally mutated genomes to daughter cells |
| 12. Melatonin | Anti-clastogenic, anti-mutagenic, anti-carcinogenic and anti-oxidant compound. |
10.10.2. Transcriptome Preserving and Repair Networks
The human transcriptome, being subjected to the same mutational events that can alter and damage the DNA, seems to have efficient anti-mutation mechanisms to guard against occurrence of errors during RNA transcription and to correct and repair some post-transcription defects of mRNA that can cause errors during protein translation. RNA editing refers to molecular modifications of nucleotides of RNA through chemical changes in the base makeup of the molecule. It involves both coding and non-coding RNA classes, and is mediated by a complex repair system comprising many species of small RNA (guide RNA) and large protein complexes known as the editosomes. Specific endonucleases and ligases for double stranded species of RNA have been defined in many prokaryotes. The pathways of RNA editing include many diverse processes: nucleoside base modifications, deamination, as well as non-templated insertions of nucleotide57. RNA editing in mRNAs effectively alters the amino acid sequence of the encoded protein so that it differs from that predicted by the genomic DNA sequence. Though mRNA editing is used in many instances to allow for synthesis by the gene of more than one protein from the same mRNA transcript, it can also be used to repair missense or termination mutations of the molecule which can have deleterious effects on the synthesized protein.
Hypotheses assuming the existence of a separate RNA-proofreading system in eukaryotes seem reasonable in view of the vital role played by proper transcription in ensuring precise translation and preservation of the identity of the proteome. This presumed system probably acts during transcription by relying on the sequence complementarity information or database stored within the complementary silent or non-transcribing strand of DNA. Sole dependence on the sequence of the active coding strand to ensure accurate transcription of a complementary mRNA might result in improper transcription if mismatch errors occur due to, e.g. polymerase dysfunction. This assumption might, partly, explain the ambiguity of some of the basic dynamic steps of gene function where seemingly needless energy is consumed for transcribing a complementary, rather than an identical, mRNA that has to be decoded again by rRNA and tRNA in the ribosome during translation. Synthesis of a complementary mRNA allows for post-transcription proofreading and repair of resulting errors. Though the existence of a pre-translation proofreading system of final mRNA would be an efficient complementary protective mechanism against improper decoding and synthesis of defective proteins, no evidence of presence of such a protective mechanism exists yet.
10.10.3. Proteome Preserving and Repair Networks
Proteins comprise the major components of all kinds of biological networks in living cells. Hence, qualitative and/or quantitative proteome defects constitute the main source of pathophysiological alterations that result upon exposure to damaging environmental factors, including mutagens, as well as to harmful intrinsic by-products of life activities within the cell. Proteome defects result in functional deterioration of biological networks with consequent pathogenesis of genetic and non-genetic disorders. Accordingly, protection of the proteome against spontaneous and induced structural defects represents a vital aspect of life preserving mechanisms of the cell. In parallel with the existence of genome and transcriptome preserving networks, proteome preserving networks responsible for repair of structural configuration defects of synthesized proteins do exist to guarantee proper post-translation modifications of proteins. However, these proteome preserving networks do not have the same priority considerations like genome and transcriptome preserving networks, as proteins are synthesized by genes and defective or deficient proteins can be replaced by normal proteins as long as their coding genes are intact. Proteome preserving and repair networks in living cells comprise a wide variety of proteins, collectively referred to as chaperones, in addition to many other catalytic proteins, or enzymes, that perform important roles either singly or as assistant factors to chaperones.
Molecular Chaperones
Chaperones are proteins that assist the covalent folding or unfolding and the assembly or disassembly of newly synthesized proteins. Chaperones are concerned primarily with protein folding and assembly of oligomeric structures. One major function of chaperones is to prevent both newly synthesized polypeptide chains and assembled subunits from being misfolded or aggregated into nonfunctional structures. This role is crucial for mediating and attaining proper post-translation structural modifications of proteins to protect the cell against pathological consequences resulting from accumulation of unfolded and misfolded proteins. Although the majority of newly synthesized proteins can fold spontaneously in a correct manner, many can not and need the assistance of specific chaperones to undergo proper folding and proper steric conformational assembly (Figure 27, Figure 28).
Figure 27.Molecular chaperones and proteome repair networks
Figure 28.Role of molecular chaperones in protein folding
There are many different families of chaperones (Hsp60, Hsp70, Hsp90, etc), each composed of large numbers of proteins. These proteins constitute the chaperone networks that exert their protective roles in protein folding and other related cellular functions as well as other critical physiological processes in the cell. Many chaperones are heat shock proteins (Hsp) because they are expressed in response to thermal stress, like elevated temperature. The reason for this behavior is that protein folding is a sensitive dynamic structural alteration that gets severely affected by heat and, therefore, many chaperones are synthesized to prevent and/or correct structural alterations caused by heat-induced misfolding. Defective protein folding induced by high temperature might be attributed to the loss of the thin water envelope that surrounds the protein molecule. Once this water layer, necessary for maintaining efficient dynamics of the protein molecule, is lost or diminished upon exposure to high temperature, defective structural configuration of the molecule, including improper folding, ensues. Other chaperones are involved in the folding of newly synthesized proteins as they are extruded from the ribosomes. Although most newly synthesized proteins can fold in absence of chaperones, many of them require chaperones for their proper folding 58.
A considerable number of chaperone proteins comprising chaperone networks function as enzymes, e.g., protein disulfide isomerase (PDI) and peptidyl prolyl cis-trans-isomerase (PPI) which act as foldases, or enzymes mediating folding of newly synthesized proteins. Protein folding by chaperone networks involves the participation of the lectin chaperones, calreticulin and calnexin, with the enzyme protein disulfide-isomerase A3, localized to the endoplasmic reticulum, to form complexes that mediate protein folding by promoting the formation of disulfide bonds in their glycoprotein substrates. In a complimentary way, other chaperones work as holdases, or enzymes that bind folding domains of newly synthesized proteins to prevent their aggregation, for example DnaJ or Hsp33 59.
Some chaperone networks within the endoplasmic reticulum function as quality control systems ensuring that only properly folded proteins are exported and secreted before being trafficked and targeted to their final cellular locations. These networks comprise different chaperone molecules like the lectin chaperones calnexin and calreticulin, N-glycan-modifying enzymes and ERp57, acting in concert. For example, stepwise cooperative functioning of these chaperones are necessary for folding newly synthesized MHC class I α-chains, which is a prerequisite step for preparing it to function properly as an antigen binding molecule 60.
Other important functions of chaperons include assistance in protein degradation, transport of proteins across membranes of cellular compartments like the mitochondria and the endoplasmic reticulum, and bacterial adhesion activity. Functional defects of many chaperones have been implicated in pathogenesis of diseases linked to protein aggregation defects like Alzheimer disease, Parkinson disease and prion diseases Winklhofer and Tatzelt 2006. Additionally, some chaperones might function as transcription regulator molecules. For instance, calreticulin can inhibit androgen receptor and retinoic acid receptor transcriptional activities in vivo, as well as retinoic acid-induced neuronal differentiation, thus acting as an important modulator of the regulation of gene transcription by nuclear hormone receptors 61.
10.11. Neuronal Brain Networks
Life activities of humans, and of other animal species, are preserved by prudent and accurate cooperative integration of functions of all cells/tissues/organs of the body. This precise integration is basically governed, conducted and maintained by the harmonized functions of the two dominating regulatory systems of the body: the nervous system and the endocrinal system. Functional analysis of the dynamics and mechanisms of action of the large number of biological networks in living cells reveals that the vast majority of these network systems obey basic physical principles of biological thermodynamics as regards reaction dynamics, energy transfer and transformation, degree of entropy, stability of components and total network performance. The networks responsible for conducting endocrinal regulatory functions act in relatively simple and direct modes.
Hormones secreted by endocrine glands either act as direct effectors of specific physiological functions or as signal transducers leading to initiation of functions of metabolic networks and/or signaling pathways. Few hormones like steroids and thyroid hormones, however, exert direct transcriptional effects, either as activators or silencers, on expression of genes encoding products necessary for executing their physiological functions.
Analysis of structural and functional organization of the human nervous system, and those of other higher order creatures, reveals that neurological functions aim at regulating three main aspects of biological life of living organisms. First, regulating and maintaining vital functions essential for preserving life, e.g., energy production, respiration, circulation, water and electrolyte balance and critical metabolic processes. Second, regulating non-essential, albeit important, functions like vision, hearing and voluntary muscle movement. Third, formulation, developing, sustaining and regulation of human-specific higher intellectual functions.
Neurological control and regulation of vital as well as of non-essential life activities happen either directly through brain centers responsible for these functions or indirectly. Direct regulation is exerted via specialized neuronal networks comprising structural components including nuclei in the brain and brainstem and other brain areas/cranial and extra-cranial nerves and intra and inter-hemisherical connections, e.g., cranial nerve nuclei present in the midbrain, pons, medulla oblongata and spinal cord/the respiratory center located in the medulla oblongata and pons/the osmoreceptors located in the supraoptic and paraventricular nuclei of the hypothalamus and the thirst center situated in the organum vasculosum of the anterior hypothalamus and glucoreceptors distributed in diffuse scattered areas of the brain 62. Indirect neurological regulation of life activities is mediated by what can be referred to as neuro-endocrinal regulatory networks located within the hypothalamus and the anterior and posterior pituitary gland. These networks comprise specialized neuronal collections and endocrinal secretory cells controlled by regulatory hormones synthesized and released by the parvo-cellular neuro-secretory cells in the hypothalamus. Controlled secretion of releasing or trophic hormones from the hypothalamus and the pituitary gland regulates hormonal secretion of all other endocrine glands in the body leading to regulation of nearly all metabolic and a wide spectrum of non-metabolic processes in body cells and organs 63.
Contrary to this simple functional design of the hormonal regulatory network systems, neural regulatory networks of the human brain act within a complex multi-scale hierarchical organization design. The organization of the human brain comprises two major distinct, though strictly tightly and precisely inter-dependent and inter-connected systems, viz. the structural and the functional systems. Application of basic principles of the graph theory to the organization design of the human brain clearly shows that it has structural and dynamic features characteristic of complex network systems 64.
This complexity might be attributed to many factors headed by the fact that in addition to its dominating regulatory role of functions of other vital organs, the human brain has to develop and sustain higher intellectual functions dependent, in essence, on its ability to gain knowledge, to process this knowledge in rational ways, and to make use of it for his welfare. Although most higher animal species, and even lower species of simple creatures, appear to share humans this ability to gain knowledge and to use it for their own sake, they are driven and dominated solely by their innate instincts. Human brain has many disjunctive peculiar features that enable it, in still unexplained mysterious ways, to gain and to acquire human-specific intellectual functions like cognition/reasoning/learning/reading and writing.
The precise underlying etiologies of these unique human cognitive capacities are, still, quite far from being elucidated or understood. Many observations, however, do exist that might be helpful in revealing the nature of some of these etiologies. For instance, it has been found that comparative study of the transcriptomes of human, chimpanzee, and macaque telencephalon revealed a number of significant findings: 1. predominance of genes differentially expressed within human frontal lobe and a striking increase in transcriptional complexity specific to the human lineage in the frontal lobe. 2. highly conserved gene expression pattern of the caudate nucleus. 3. gene co-expression signatures related to FOXP2 gene which is important for language evolution 65. Also, it has been found that attention, which constitutes a fundamental characteristic of human cognition, is dependent on presence and development of human-specific attention neural networks some of which are lacking in higher close animal species 66.
Unique, cytological and histological features of human brain cells have been observed for many cell types. Astrocytes of human brain, for instance, have substantially peculiar morphological features and important functional capabilities including remarkable large size, possession of dendritic extensions that penetrate deeply through several layers of cortical grey matter, intimate vicinity to large numbers of neurons and synapses, high-speed communication with other adjacent astrocytes and neurons through inter-cellular networks using calcium signaling as neurotransmitter pathway, regulation of intra and inter-neuronal ion flux and neurotransmitter concentrations necessary for proper neuronal-synaptic communications, and secretion of other important neuro-transmitters, e.g., TNFalpha, that enhance synaptic transmission. These observations strongly suggest crucial regulatory roles played by glia cells of the brain, mainly astrocytes, in processing of information and in modifying transmission and flow of information through specific neuronal networks responsible for development of higher intellectual functions like memory, learning, cognition and intelligence 67.
The nature of neuronal brain network systems responsible for formulation, development and preservation of higher intellectual functions remains, largely, unknown. The extreme complexity of the fine structural design of the human brain represent a major obstacle as accurately attributing specific cognitive and intellectual functions to particular collections of neurons, among an estimated 100 billion neurons, of the brain is nearly an unimaginable task. Also, correlations between specific higher intellectual functions and particular regions of the brain based on findings observed in experimental animals can not be applied for human brains, because counterparts of many human-specific intellectual and emotional responses to various internal and external stimuli could never be defined for, or detected in, or expressed by experimental animals. In addition, application of observations and findings in experimental animals to human brains can never be justified, except for biochemical changes and physiological alterations, in view of the marked structural and functional differences between human brain and brains of even the nearest phylogenetically related higher animals.
Though postulations attributing major effective roles to the proteome of the neuron, the neuro-proteome, as final mediator of these functions seem reasonable upon comparison with similar roles played by the proteome of cells of other organs, the roles played by the genome of the neuron, the neuro-genome, in appearance and development of these functions are, still, quite vague and unexplainable. Many different components of the neuro-proteome have been related to, and implicated in, development of different intellectual functions. Prion proteins, for instance, have been hypothesized to be correlated with memory or storage and recall of information 68, and prion-like proteins have been postulated to be responsible for development of long-term memories in mice, and probably in other mammals 69. A global correlation between proteins and memory has been inferred from the observation that inhibition of protein synthesis leads to weakening, or even loss, of memory 70. Tens of different proteins with varying functions, e.g., DNA-binding proteins and enzymes involved in DNA repair, have been postulated to play roles in memory processes based on experimental findings in knockout mouse. In spite of these theoretical postulations deduced from experimental work on animals, their significance for human memory can never be validated, and a causal role of studied proteins in development of memory or other cognitive functions has not yet been established.
Since proteins are the direct effective mediators of all life activities in living cells whether acting as catalytic/ structural/signaling or regulatory biomolecules, hypotheses concerning their presumed roles in development and execution of intellectual brain functions seem quite reasonable. However, refining some of the basic postulations of these hypotheses is mandatory in view of the vague mechanisms of action attributed to proteins in this respect. The statement that proteins can store information is exceedingly puzzling and hard to conceive for many reasons.
First: there is no single pattern of information input to neurons, some data are perceived as electric impulses, some data are caused by, and recognized as, biochemical alterations, and some data are generated in response to endogenous and/or external physical changes. Still, the sources and nature of responses concerning the unexplainable features and the characteristic mysterious aspects of human behavior and demeanor, like emotions/imaginations/affections, are hard or even impossible to explain within materialistic contexts of laws of physics and chemistry.
Second: the instantaneous nature of cognitive or intellectual responses of the brain to some peculiar forms of external stimuli, visual or auditory stimuli for instance, can not be accounted for by the concomitant dynamic changes in protein conformation presumed to be responsible for eliciting/recalling/interpreting or realization of these responses. This assumption might be applicable for stimuli elicited by biochemical changes inside or outside the neuron in response to certain types of stimuli, e.g., metabolic alterations. Dynamic changes in protein conformation are, relatively, time consuming processes and are hard to accept as sole effectors of instantaneous cognitive or impulsive responses to many types of external stimuli that need multi-stage processing mechanisms before their conveyance, perception and interpretation by the brain.
Third: protein denaturation, a continuously occurring deleterious structural conformational change of proteins inside all living cells, in response to detrimental damaging factors like oxidative stress/radiation induced damage/metabolic alterations etc., can explain, at least in part, many abnormal states of intellectual impairment, cognitive regression, and memory defects like forgetting/amnesia/memory distortion. Prompt degradation of denatured proteins to their constituent amino acids by specific autolytic enzyme systems in the cell occurs leading to selective deficiencies of denatured proteins. However, in normal healthy conditions, balanced compensatory replacement of deficient or defective proteins by the genome restores and maintains the integrity and efficiency of these higher mental processes. Occurrence or persistence of these abnormalities in absence of underlying genetic defects or absence of evidence of deficient compensatory replacement of degraded proteins, however, points to possible participation and involvement of other factors in mechanisms responsible for storage and processing of information by the brain.
11. Proposed Hypothesis of Mechanisms of Function of Neuronal Networks
In view of the characteristic features and distinctive nature of the wide spectrum of structural and functional properties of the nervous system of higher organisms, in general, and of the human brain in particular, few apparently plausible hypotheses regarding mechanisms of development and mediation of higher intellectual functions in human brain might be proposed as follows:
1. Perception, propagation and flow of information, gained from endogenous sources and external stimuli, to neurons occur first as specific defined patterns of electric impulses. Metabolic changes in the extra-neuronal environment might, also, be conveyed to the inside of the neuron first as electrical input to be translated to specific proteomic responses via signaling networks within the neuron.
2. Dendritic input to the neuron might elicit, through certain signaling pathways, specific proteomic responses that functionally relate particular proteins to specific patterns of information input. Contrary to the majority of physiological processes mediated by the activity of single enzymes, or few enzymes and protein molecules, the complex and intricate nature of mental processes make them difficult to be executed by single or few proteins and considerable number of proteins might be involved in these processes.
3. Recognition of specific patterns of information input may cause relevant specific neuronal network(s), composed of afferent nerves, dendrites, signaling pathways, and particular arrays of certain proteins, to get into work and a specific imprint formulated by the combined electric and proteomic responses to information input of particular situations is constructed and preserved.
4. The huge, unimaginable quantitative and qualitative spectra of information flow and input to the brain necessitates involvement and participation of multiple specific information imprints, each devoted for specific input state, in formulation of other particular imprints for other different situations. Thus, few or variable numbers of imprints, where each is peculiar for a specific input state, might be collectively and/or selectively combined to formulate and characterize a different mental, cognitive or intellectual condition.
5. This presumed team work design of neuronal functions postulates presence of functional specialization of the large numbers of neurons constituting particular components of the brain. A structural-functional design of the human brain according to this assumption is built on the postulation that within each cellular neuronal component of the brain, e.g., the cerebral cortex/nuclei of cranial nerves/the amygdala/the thalamus/the hypothalamus etc., functionally predefined groups of neurons do exist. Each functionally specialized neuronal group probably works on network basis, i.e. they form a well- defined structured functional network composed of the neurons and their functionally related nerve fibers/dendritic processes and ramifications/support and regulatory glial cells and, probably, other poorly defined regulatory or signaling pathways necessary for combining/synchronizing and harmonizing their functions with other groups.
6. Perception, storage and archiving of different types of information with formulation of specific functional imprints and specialized networks, each being selectively responsible for handling one or more distinctive type of information input to the neuronal group(s), might represent the afferent limb of information storing mechanism(s) of the brain. The exceedingly large number of neurons composing the human brain, estimated to count around one billion neurons, and the much more extensive inter-neuronal connections amounting to more than one trillion connections, allow for efficient handling, processing and storing of the unimaginable flow of daily, sometimes hourly, information input and flow to the human brain. Theoretically, millions and even billions of information-handling specialized neuronal networks with comparable numbers of specific imprints for all possible kinds of information-processing mental operations confronted with in daily life, could be constructed and formulated in the human brain in view of the huge numbers of neurons and inter-neuronal connections composing the nervous system.
7. According to these preliminary hypotheses, storing/processing/archiving of information in the neuron seem to be a dynamic conditioned process controlled by specialized neuronal networks comprising proteins as one of their components, rather than a static process depending on lasting presence of certain proteins specialized in storing information. The network concept, instead of the protein concept, as regards information handling by the neuron seems more palatable for many reasonable considerations:
a. The number of proteins needed to execute different stages of information handling, e.g., perception/recognition/archiving/formulation of specific imprints/storage etc, by the neuron would be huge enough to cause considerable spatial and functional encroachment on other neuronal functions needed to sustain its essential life activities.
b. A sizable portion of the genome would be needed and devoted for synthesis of required proteins with consequent deleterious effects on synthesis of proteins mediating other life activities of the cell.
c. The multistage dynamic nature of information handling cascade inside the neuron necessitates and imposes many dynamic communications and mutual interactions between many different related components, e.g., proteins/signaling molecules/regulatory molecules/modulating pathways etc. Obviously, efficient and proper execution of these stages and processes could only be mediated and performed according to basic rules and mechanisms of network functioning. Attributing the ability to execute these mechanisms to structured designs of static proteins, whether in groups or in scattered patterns in the cytosol, seems unreasonable as it contradicts the basic principles of physics, chemistry and efficient energy consumption rules of thermodynamics.
8. Hypotheses regarding the nature of mechanisms implicated in, and responsible for, recall and retrieval of information imprints stored within the neurons are much more difficult to formulate. Normal physiological and pathological interpretations of mechanisms of neuronal control of other body organs in health and disease, respectively, provide relatively acceptable explanations of the nature of neuronal responses and brain function within the context of known functional mechanisms of the nervous system. Neurological regulation of somatic and autonomic body functions is exerted by many different mechanisms including, for example, autonomic and somatic reflex arcs initiated and conducted by monosynaptic and polysynaptic pathways, feedback potentiation or inhibition of afferent or efferent pathways, neuro-endocrinal regulation of secretory functions of endocrine glands, and the like. Unfortunately, no comparable assumptions as regards possible mechanisms involved in storage, processing, recall and retrieval of stored information generated by higher cognitive and intellectual functions, e.g., recognition, intuition, thinking, reasoning talking, learning, writing, memory, emotions and imagination, could be speculated or formulated. In spite of the currently available flood of information regarding the fine structure of the nervous system, and to a lesser extent, its functional capabilities, sufficient knowledge of the structural and functional designs of the brain at the molecular level is grossly lacking. Unavailability of more specific non-invasive diagnostic techniques and analytical approaches to disclose these mysterious aspects of the human brain at more deeper insights represents a major obstacle in this respect. It is hoped that disclosure of intra-neuronal structural and functional specialization of the neuro-proteome, and possible network systems responsible for higher functions of the brain, would add great wealth of data indispensable for formulating better hypotheses regarding the nature and sources of these functions. It would, also, be of great help in postulating better assumptions regarding the construction, development, and the mechanisms of action(s) of neuronal brain networks responsible for intellectual functions of the brain.
9. The abovementioned hypothetical postulations regarding possible mechanisms of information processing in the brain might offer a partial interpretation of some memory recall and retrieval defects like temporary and/or recurrent amnesia and Alzheimer disease. Degradation of key proteins that participate in neuronal networks responsible for performing specific memory-storage and recall functions, might explain temporary amnesia due to interruption with function of these networks. Re-synthesis and replacement of degraded/denatured/deficient proteins might, in a similar way, explain regaining lost functions related to memory recall and retrieval. However, if considerable loss of functions of genes responsible for synthesis of these proteins, e.g., due to un-repairable mutation-induced damage, happens, permanent loss of memory for many events, depending on the magnitude and the spectrum of these events, may occur.
12. Challenges to Defining Key Regulatory Molecules of Biological Networks
Defining key regulatory molecules of networks implicated in pathogenesis and progression of the majority of genetic, as well as most non-genetic, diseases lacking obvious clues as regards possible underlying etiology, represents a hard task and a real challenge because of many difficulties including (Table 5):
1. The exceedingly large number of biological networks within the cell.
2. The ever changing dynamic frame of interactions/interconnections and communications between large numbers of functionally related networks within the cell and subcellular compartments.
3. The complex nature of networks that control and synchronize the functional performance of the tremendous pathways mediating intercellular/inter-tissues/inter-organs connections and behavior.
4. The, still, undisclosed nature and designs of the mysterious genetically-determined master network systems responsible for regulating and harmonizing functions of all other biological networks of all body components.
5. The enigmatic nature and the, largely, unknown codes and rules of the predefined and pre-programmed frame of life that guarantees fulfillment of the essential requirements of optimal biological fitness of living creatures.
Table 5. Challenges to defining key regulatory molecules of biological networks| 1. Exceedingly large number of biological networks within the cell and sub-cellular components. |
| 2. Continuously changing dynamic reactions/interactions of network components. |
| 3. Multi-functionality of structural/catalytic components of many related/unrelated networks. |
| 4. Undisclosed nature and designs of genetically-determined master regulatory network systems. |
| 5. Enigmatic nature of codes and rules of predefined and pre-programmed frame of life. |
13. Importance of Studying and Analysis of Biological Networks
Meticulous study of biological networks represents an important and integral aspect in study of biology. Interpretation and analysis of basic information deduced from observing and analyzing structural designs and functional characteristics and dynamics of biological networks discloses and defines the basic framework within which life activities in living cells are initiated, adapted to physiological requirements, maintained, and terminated upon completion of their aims. More important, however, is the contribution of this information to proper understanding of the different mechanisms responsible for regulating and synchronizing the functions and performances of the vast spectrum of different network categories within the cell.
Study and analysis of the structural designs and the functional mechanisms of biological networks would have crucial and important impacts on many theoretical and applied aspects of biology, in general, and of medical sciences in particular. As regards medical genetics, these aspects include many diagnostic, therapeutic, prophylactic, biotechnological and theoretical applications. For example, discovering and defining the key pivotal structural and regulatory molecules within life-mediating networks, and along different pathways responsible for controlling functional dynamics of the network, represent an indispensable approach insistent for designing proper therapeutic approaches to diseases caused by network defects. This approach has been successfully adopted and resorted to since decades for treatment of many genetic diseases, e.g., enzyme replacement therapy for metabolic errors due to enzyme deficiencies and protein replacement therapy for protein deficiency disorders like immunodeficiency and hemophilia.
In a parallel way, defining key regulatory molecules of biological networks in normal conditions would have a pivotal impact on diagnostic approaches to diseases caused by qualitative and/or quantitative defects of these molecules. For instance, comparative analysis of normal profiles of these molecules with their profiles in specific disease conditions can point out defects leading to qualitative and/or quantitative abnormalities in their synthesis. Furthermore, in a way similar to the concept of reverse genetic engineering, categories of RNAs involved in translation and synthesis of these molecules and even abnormal/mutated genes involved in coding and defining their structural designs and functional roles can be detected and defined in ambiguous disease states caused by defective proteins coded by, still, undefined genes.
Within a global context, success in defining the whole proteome spectrum of specific cells would have a great and crucial impact on both diagnostic and therapeutic approaches to some dreadful human diseases that represent real challenges to researches aiming at their early diagnosis and effective treatment. If this goal can be achieved, early and accurate diagnosis of specific cancers, for instance, might be possible upon comparing the whole proteome of suspected malignant cells with their counterpart of normal cells. Comparative analysis of observed deviations of malignant proteomes might even disclose unapparent aspects of tumor progression and ongoing changes of the malignant phenotype more accurately and at much earlier stages than can be achieved by currently available traditional diagnostic techniques. This same approach might prove useful for other mystical disease conditions including, for instance, many neurological and autoimmune disorders.
Defining and characterizing biological networks and biochemical pathways responsible for mediating specific stages of embryonic and fetal development can have a great impact on prophylactic approaches aiming at obviating pathogenesis of congenital abnormalities caused by defective or deficient synthesis of structural proteins/catalytic enzymes of these networks and pathways. Achieving this goal necessitates, firstly, construction of normal developmental proteome maps composed by complete and accurate characterization of the structural components of the vast majority of these regulatory networks and pathways at different temporal stages of growth and development for different tissues and organs. Comparing databases of these normal developmental proteome maps with their counterparts of embryonic or fetal developmental maps for the same temporal stage of development of specific tissues or organs can offer an early and accurate diagnostic, and possibly prophylactic, approach to structural and/or functional congenital defects of tissues or organs with abnormal and deviant developmental maps. A major obstacle in this regard is the restriction imposed by the time limits that govern the transfer of embryonic/fetal cells to maternal circulation where they can be separated and analyzed.
Construction of genomic developmental maps composed of databases of expression states of genes/genome parts during specific temporal stages of development might prove to be the ultimate proper and effective approach for proper and accurate preparation/selection/engineering of specific stem cells suitable for replacement therapy of diseased tissues or organs in post-natal life. The genomes of stem cells are strictly organized under extreme and strict programming rules that regulate and define their final expression states at time of their formation during embryonic/fetal life and during their latency in post-natal life, as well. The specific genome of a specific stem cell comprises thousands of active genes and similar number of temporally, and spatially, imprinted inactive genes. This state of pre-programmed and pre-defined selective activation and selective repression of the genomes of stem cells occurs only within an exquisite environment comprising the actions of thousands of genes of the genome and their interactions with similar number of genes of other effector or regulatory genomes at specific times of growth and development, a situation nearly impossible to imitate in vitro or even in vivo in post-natal life. Proposed approaches to construct genomic developmental maps might prove useful in re-engineering the genomes of stem cells to render them capable of re-performing their roles during development in post-natal life.
Hypothetical aspects regarding possible impacts of studying biological networks on eugenics might become a field of research and experimentation in the near future. For instance, theoretical proposals aiming at improving human capabilities, fitness, and adaptation to drastic environmental conditions by genetic engineering techniques that can improve the performance and efficiency of native genomic and proteomic networks of human beings, can be formulated and rendered applicable for in vitro then for in vivo experimentation. Researches in this regard might be directed basically towards improving somatic characteristics of human beings via engineering human genome and proteome to acquire characteristics of genomes and proteomes of animals that have better somatic features and capabilities than human beings. For example, reengineering human proteome networks responsible for hearing or vision or smelling to mimic those of animals characterized by supreme capabilities of these senses might be tried, in spite of the short term eugenic consequences of such trials as they are limited by the short life span and relatively rapid turnover rates of engineered protein components of the proteome networks. Trials aiming at reengineering proteome networks, or genome networks, of human beings for eugenic purposes might seem attractive for many obvious reasons, but they implicate considerable risks that can neither be ignored nor tolerated for ethical, health, and medical reasons in view of the unique identity of the human race compared with identities of all other species. In view of the current state of vague and scanty knowledge available as regards the structural/functional/regulatory aspects of the human genome and human proteome, researches directed to disclose details of these aspects would represent the proper attitude towards defining the best diagnostic, therapeutic, and prophylactic approaches for human diseases.
References
- 1.Gyurkó D, Dániel V, Dezső M, Katalin L, Tamás K. (2013) Adaptation and learning of molecular networks as a description of cancer development at the systems-level: potential use in anti-cancer therapies. Seminars in Cancer Biology. 23(4), 262-269.
- 2.Piatigorsky Joram.Gene sharing and evolution: The diversity of protein functions. (2007).Harvard University Press.P. 320.
- 3.H E Daphne, Huberts W, Klei Ida J van der. (2010) Moonlighting proteins: An intriguing mode of multitasking. , Biochimica et Biophysica Acta 1803(4), 520-525.
- 4.Zhao Renbin, Gish Kurt, Murphy Maureen, Yin Yuxin, Notterman Daniel. (2000) Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. , Genes & Development 14, 981-993.
- 5.Laggerbauer Bernhard.Dirk Ostareck, Eva-Maria Keidel, Antje Ostareck-Lederer, Utz Fischer.(2011)Evidence that fragile X mental retardation protein is a negative regulator of translation. , Hum. Mol. Genet 10(4), 329-338.
- 6.Radtke F, Georgiev O, H P Müller, Brugnera E, Schaffner W. (1995) Functional domains of the heavy metal-responsive transcription regulator MTF-1. Nucleic Acids Res. 23(12), 2277-2286.
- 7.Maumita M, Ronald B. (2004) Gene regulation by riboswitches. , Nature Reviews Molecular Cell Biology 5, 451-463.
- 8.Serganov Alexander, Pate Dinshaw. (2007) Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. , Nature Reviews Genetics 8, 776-790.
- 9.J M Vaquerizas, S K Kummerfeld, S A Teichmann, N M Luscombe. (2009) A census of human transcription factors: function, expression and evolution. , Nature Reviews Genetics 10(4), 252-263.
- 10.Hu Xiao-Pan, Yang Yi, Ma Bin-Guang. (2015) Amino acid flux from metabolic network benefits protein translation: the role of resource availability. Scientific Reports. 5, 11113.
- 11.S E Voiculescu, Zygouropoulos N, C D Zahiu, A M Zagrean. (2014) Role of melatonin in embryo/fetal development. , J. Med Life 7(4), 488-492.
- 12.A E Bely, K G Nyberg. (2009) Evolution of animal regeneration: re-emergence of a field. , Trends in Ecology & Evolution 25, 161-170.
- 13.A W Seifert, S G Kiama, M G Seifert, J R Goheen, T M Palmer. (2012) Skin shedding and tissue regeneration in African spiny mice (Acomys). , Nature 489(7417), 561-565.
- 14.J A Baddour, Sousounis K, P A Tsonis. (2012) Organ repair and regeneration: an overview. Birth Defects Res. 96(1), 1-29.
- 15.R T Böttcher, Niehrs C. (2005) Fibroblast growth factor signaling during early vertebrate development. Endocrine Reviews. 26(1), 63-77.
- 16.O S Soyer, Salathé M, Bonhoeffer S. (2005) Signal transduction networks: topology, response and biochemical processes. , J Theor Biol 238(2), 416-25.
- 17.David A Fruman, Craig M Walsh. (2007) Signal transduction and autoimmunity:. , Introduction. Autoimmunity 40, 403-404.
- 18.Eric K Rowinsky. (2003) Signal Events: Cell signal transduction and its inhibition in cancer. The Oncologist;8(Suppl.3):. 5-17.
- 19.Kathryn R Bowles, Jones Lesley. (2014) Kinase Signalling in Huntington’s Disease. , Journal of Huntington’s Disease 3, 89-123.
- 20.C A Anastassiou, Perin R, Markram H, Koch C. (2011) Ephaptic coupling of cortical neurons. , Nature Neuroscience 14(2), 217-223.
- 21.Ebrey T, Koutalos Y. (2001) Vertebrate photoreceptors. Progress in Retinal and Eye Research 20(1), 49-94.
- 22.Komiya Yuko, Habas Raymond. (2008) Wnt signal transduction pathways. , Organogenesis 4(2), 68-75.
- 24.Choudhry Zia.Azadeh A Rikani, Adnan Maqsood Choudhry, Sadaf Tariq, Fozia Zakaria, et al (2014) Sonic hedgehog signalling pathway: a complex network. , Ann Neurosci 21(1), 28-31.
- 25.A M Skoda, Simovic D, Karin V, Kardum V, Vranic S. (2018) The role of the Hedgehog signaling pathway in cancer: A comprehensive review. , Bosn J Basic Med Sci 18(1), 8-20.
- 26.Liu Hailan, Gu Dongsheng, Xie Jingwu. (2011) Clinical implications of hedgehog signaling pathway inhibitors. , Chin J Cancer 30(1), 13-26.
- 27.Heussler H S, Suri M, I D Young, Muenke M. (2002) Extreme variability of expression of a Sonic Hedgehog mutation: attention difficulties and holoprosencephaly. Archives of disease in childhood. 86, 293-296.
- 28.Surabhi S, B K Tripathi, Maurya B, P K Bhaskar, Mukherjee A. (2015) Regulation of Notch signaling by an evolutionary conserved DEAD Box RNA Helicase, Maheshvara in Drosophila melanogaster. , Genetics 201(3), 1071-1085.
- 30.Peter D Turnpenny. (2014) . Syndromes and Diseases Associated with the Notch Signaling Pathway. Doi:org/10.1002/9780470015902.a0024870 .
- 31.Horbelt D, Denkis A, Knaus P. (2012) A portrait of Transforming Growth Factor β superfamily signalling: Background matters. , Int. J. Biochem. Cell Biol 44(3), 469-74.
- 32.H C Patterson, Gerbeth C, Thiru P, N F Vögtle, Knoll M. (2015) A respiratory chain controlled signal transduction cascade in the mitochondrial inter-membrane space mediates hydrogen peroxide signaling. , PNAS 112(42), 5679-88.
- 33.Alexandra M Fajardo, Gary A Piazza, Heather N Tinsley. (2014) The role of cyclic nucleotide signaling pathways in cancer: targets for prevention and treatment. Cancers (Basel). 6(1), 436-458.
- 34.Matthias P Wymann, Schneiter Roger. (2008) Lipid signalling in disease. , Nature Reviews Molecular Cell Biol 9, 162-176.
- 35.M Y Leo, Leung Chun-Yin, Walfred W C, Choi Heung-Ling, Leung Yun-Chung. (2012) A paradoxical teratogenic mechanism for retinoic acid. , PNAS 109(34), 13668-13673.
- 36.Bhaskar C Das, Thapa Pritam, Karki Radha, Das Sasmita, Mahapatra Sweta. (2014) Retinoic acid signaling pathways in development and diseases. , Bioorg Med Chem;15: 22(2), 673-683.
- 37.P J Murray. (2007) The JAK-STAT signaling pathway: input and output integration. , J 178(5), 2623-2629.
- 38.Pencik J, Pham H T, Schmoellerl J, Javaheri T, Schlederer M. (2016) JAK-STAT signaling in cancer: From cytokines to non-coding genome. , Cytokine 87, 26-36.
- 39.Haixia C, Renpeng G, Qian Z, Hongchao G, Meng Y. (2015) Erk signaling is indispensable for genomic stability and self-renewal of mouse embryonic stem cells. , PNAS 112(44), 5936-5943.
- 40.Laplante M, D M Sabatini. (2012) mTOR signaling in growth control and disease. , Cell 149(2), 274-293.
- 41.Tang G, Gudsnuk K, S H Kuo, M L Cotrina, Rosoklija G. (2014) Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. , Neuron 83, 1131-1143.
- 43.Xu K, Liu P, Wei W. (2014) mTOR signaling in tumorigenesis. , Biochimica Et Biophysica Acta 1846(2), 638-54.
- 44.Pópulo H, J M Lopes, Soares P. (2012) The mTOR signaling pathway in human cancer. , International Journal of Molecular Sciences 13(2), 1886-1918.
- 45.Faivre S, Kroemer G, Raymond E. (2006) Current development of mTOR inhibitors as anticancer agents. Nat Rev Drug Discov. 5(8), 671-88.
- 46.Saucedo Leslie J, Bruce A Edgar. (2007) Filling Out the Hippo Pathway. Nature Reviews Molecular Cell Biology. 8(8), 613-621.
- 47.M J Berridge, Peter Lipp, Bootman M D. (2000) The versatility and universality of calcium signaling. , Nature Reviews Molecular Cell Biology 1(1), 11-21.
- 48.Elmore Susan. (2007) Apoptosis: a review of programmed cell death. , Toxicol Pathol 35(4), 495-516.
- 49.S E Logue, S J Martin. (2008) Caspase activation cascades in apoptosis. Biochem Soc Trans.;36(Pt.1):. 1-9.
- 50.Paula C Ashe, Mark D Berry. (2003) Apoptotic signaling cascades. Progress in Neuro-Psychopharmacology & Biological Psychiatry 27, 199-214.
- 52.Emily M, Satchidananda P. (2016) Circadian clock, nutrient quality, and eating pattern tune diurnal rhythms in the mitochondrial proteome. , PNAS 113(12), 3127-3129.
- 53.Cajochen C, Krauchi K, Wirz-Justice A. (2003) Role of Melatonin in the regulation of human circadian rhythms and sleep. , Journal of Neuroendocrinology 15, 432-437.
- 54.Ko Caroline, Takahashi Joseph. (2006) Molecular components of the mammalian circadian clock. , Hum. Mol. Genet.15(sup.2): 271-277.
- 55.Uhlhaas Peter, Singer Wolf. (2011) Developmental changes in neuronal oscillations and synchrony: evidence for a late critical period. Human Neuroplasticity and Education. 117, 218-260.
- 56.Zaslaver Alon, Mayo Avi, Rosenberg Revital, Bashkin Pnina, Sberro Hila. (2004) Just-in-time transcription program in metabolic pathways. , Nature Genetics 36, 486-491.
- 57.Takenaka M, Zehrmann A, Verbitskiy D, Hartel B, Brennicke A. (2013) RNA editing in plants and its evolution. , Annual Review of Plant Biology 47, 335-352.
- 58.R J Ellis. (2006) Molecular chaperones: assisting assembly in addition to folding. Trends in Biochemical Sciences. 31(7), 395-401.
- 59.Hoffmann J R H, Linke K, P C Graf, Lilie H, Jakob U. (2003) Identification of a redox-regulated chaperone network. , The EMBO Journal 23(1), 160-168.
- 60.Benyair R, Ron E, G Z Lederkremer. (2011) Protein quality control, retention, and degradation at the endoplasmic reticulum. , Int. Rev. Cell. Mol. Biol 292, 197-280.
- 61.J M Tarr, P J Young, Morse R, D J Shaw, Haigh R. (2010) A mechanism of release of calreticulin from cells during apoptosis. , Journal of Molecular Biology 401(5), 799-812.
- 62.Park Hae-Jeong, Friston Karl. (2013) Structural and functional brain networks: from connections to cognition. , Science 342(6158), 579.
- 63.Barrett Kim.Susan Barman, Scott Boitano, Heddwen Brooks (2012)Ganong's review of medical physiology. 24th ed. LANGE Basic Science
- 64.Bullmore Ed, Sporns Olaf. (2009) Complex brain networks: graph theoretical analysis of structural and functional systems. , Nature reviews neuroscience 10, 186-198.
- 65.Konopka G, Friedrich T, Davis-Turak J, Winden K, M C Oldham. (2012) . Human-specific transcriptional networks in the brain. Neuron.23: 75(4), 601-617.
- 66.Patel Gaurav, Yang Danica, Jamerson Emery, Snyder Lawrence, Corbetta Maurizio. (2015) Functional evolution of new and expanded attention networks in humans. , PNAS 112(30), 9454-9459.
- 67.Robertson James. (2014) Astrocytes and the evolution of the human brain. Medical hypotheses. 82(2), 236-239.
- 68.Tompa P, Friedrich P. (1998) Prion proteins as memory molecules: an hypothesis. , Neuroscience 86(4), 1037-1043.
Cited by (14)
- 1.Dobovišek Andrej, Vitas Marko, Blaževič Tina, Markovič Rene, Marhl Marko, et al, 2023, Self-Organization of Enzyme-Catalyzed Reactions Studied by the Maximum Entropy Production Principle, International Journal of Molecular Sciences, 24(10), 8734, 10.3390/ijms24108734
- 2.Arif Samia Muhammad, Mustafa Ghazala, Cheng Kejun, 2024, , , (), 189, 10.1007/978-981-97-2918-0_11
- 3.Wadood Armughan Ahmed, Xiquan Zhang, 2024, Unraveling the mysteries of chicken proteomics: Insights into follicle development and reproduction, Journal of Proteomics, 308(), 105281, 10.1016/j.jprot.2024.105281
- 4.Sasidharakurup Hemalatha, Menon Anil S., Sabeen Avinash Sreedharan, Diwakar Shyam, 2022, , , 1373(), 181, 10.1007/978-981-16-4369-9_19
- 5.Zhang Duzhen, Zhang Tielin, Jia Shuncheng, Wang Qingyu, Xu Bo, 2024, Tuning Synaptic Connections Instead of Weights by Genetic Algorithm in Spiking Policy Network, Machine Intelligence Research, 21(5), 906, 10.1007/s11633-023-1481-1
- 6.Vijitha R. Jeba, Beauno S., 2024, SOLITON EXCITATIONS IN AN (3+1) DIMENSIONAL Alpha-HELIX PROTEIN SYSTEM WITH QUADRAPOLE-QUADRAPOLE INTERACTIONS, ShodhKosh: Journal of Visual and Performing Arts, 5(6), 10.29121/shodhkosh.v5.i6.2024.3464
- 7.Lakshmi Lingidi Jhansi, Rathore Savita, Faizal Muhamed, Khan Yusuf Faiz Noor, Zephy Doddigarla, 2022, INSULIN RESISTANCE ALTER THE LEVELS OF IL-4, IL-5 AND IL-13 IN TYPE 2 DIABETES MELLITUS WITH LESS THAN 5 YEARS OF DURATION, INDIAN JOURNAL OF APPLIED RESEARCH, (), 40, 10.36106/ijar/9801433
- 8.Kumari Riya, Nag Moupriya, Lahiri Dibyajit, Bhattacharya Debasmita, Ghosh Shreosee, et al, 2025, , , 19(), 247, 10.1007/978-3-031-93189-5_10
- 9.Chakraborty Tanujit, Chattopadhyay Swarup, Das Suchismita, Naik Shraddha M., Hens Chittaranjan, 2025, A compounded Burr probability distribution for fitting heavy-tailed data with applications to biological networks, Chaos: An Interdisciplinary Journal of Nonlinear Science, 35(6), 10.1063/5.0270403
- 10.Chakraborty Sohini, Banerjee Satarupa, 2024, , , (), 195, 10.1007/978-981-99-9462-5_9
- 11.Dobovišek Andrej, Blaževič Tina, Kralj Samo, Fajmut Aleš, 2025, Enzyme cascade to enzyme complex phase-transition-like transformation studied by the maximum entropy production principle, Cell Reports Physical Science, 6(2), 102400, 10.1016/j.xcrp.2024.102400
- 12.Nguyen Trong-The, Dao Thi-Kien, Pham Duc-Tinh, Duong Thi-Hoan, 2024, Exploring the Molecular Terrain: A Survey of Analytical Methods for Biological Network Analysis, Symmetry, 16(4), 462, 10.3390/sym16040462
- 13.Solis-Perales Gualberto, Espinoza-Valdez Aurora, Luna-Oliveros Beatriz C., Rivera Jorge, Sánchez-Estrada Jairo, 2025, Design of Stable Signed Laplacian Matrices with Mixed Attractive–Repulsive Couplings for Complete In-Phase Synchronization, Mathematics, 13(17), 2741, 10.3390/math13172741