Abstract
The phytochemicals are endowed with antioxidant activities because of the presence of plentiful polyphenols and many other phytochemicals. However, some recent reports have suggested that phytochemicals from certain plant species exhibit DNA damaging properties mainly due to presence of alkaloids. In the present study, pBR322, Salmonella typhi DNA, insect DNA and human DNA were treated with hexanolic extract of Argemone mexicana and Thevesia peruviana leaves to assess their DNA damaging abilities. Another set of experiments was carried out using the methanolic extracts of Citrus lemon leaves to assess their DNA protecting abilities from damage of DNA by extracts of A. mexicana and T. peruviana at 150000 ppm for all DNAs used. The results indicated that the leaves extract of A. mexicanaand T. peruviana demonstrated significant DNA damaging potential at higher concentrations. In contrast, the extracts from C. limonat 15000 ppm showed maximum DNA protective properties for all DNAs used.
Author Contributions
Academic Editor: Wentao Xu, Food safety and molecular biology, China.
Checked for plagiarism: Yes
Review by: Single-blind
Copyright © 2020 Nitika Singh, et al.
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
The authors have declared that no competing interests exist.
Citation:
Introduction
DNA is a complex molecule that contains the information necessary to build and maintain an organism. All living things have DNA as their genetic material. It also serves as the primary unit of heredity in all the organisms. In the environment, the continuous exposure of organisms to various hazardous materials has been responsible for emergence of many genetic abnormalities 1. The abnormalities might be inherited from one generation to the other. The in vitro studies have indicated the application of Fenton reagent as a tool to demonstrate DNA damage via production of free radicals. In this reaction, hydrogen peroxide oxidizes Fe2+ to Fe3+ and produces free hydroxyl radicals (OH•) and a hydroxide ion (OH−). The hydrogen peroxide is also involved in reduction of Fe3+ to Fe2+ and generate a hydroperoxyl radical (HOO•) and a H+; shown as following:
Fe2+ + H2O2 → Fe3+ + HO• + OH−
Fe3+ + H2O2 → Fe2+ + HOO• + H+
When reactive oxygen species attack on DNA, the nitrogenous bases of DNA strands get oxidized frequently 2. The production of 8-oxoguanine or 7,8-dihydro-8-oxoguanine/8-oxo-Gua or 8-OH-Gua is the most common biomarker of various forms of oxidative DNA damage in which a specific base i.e. guanosine gets oxidized in DNA. It results into increase in the level of hydroxydeoxyguanosine (8-OHdG) 3. The 8-oxo-Gua is reported as premutagenic and also to contribute to several human diseases 4. To maintain the genetic stability from one generation to next generation, living organisms have repair systems against oxidative DNA damage 5.
The plant-based products have been reported to exhibit dual behavior such as DNA protection and DNA damage 6. The compounds present in plant extract crosslink DNA on opposite strands of the double helix (inter strand cross links), resulting in its damage. However, the antioxidants are usually reducing agents, such as vitamins, carotenoids, and polyphenols, which can scavenge reactive oxygen species and inhibit the chain reaction by donating an electron to the free radicals and neutralize them. The antioxidant defense system, supported by dietary antioxidants, protects the body from free radicals; however, during oxidative stress, antioxidants are insufficient to maintain homeostasis. The methanolic extracts of C. limon leaves have been utilized to assess their DNA protecting ability 7.
On the basis of an extensive literature survey, it has been evidenced that no research has been conducted on the DNA damaging potential of A. mexicana leaves extract though DNA damaging potential of A. mexicanafruits and seeds oil have been reported 8. While in case of T. peruviana cellular and organ toxicity has been demonstrated from the extracts of its different parts 9. The plant extract of C. limonhas antioxidant property. However, nobody has reported its DNA protective potential against oxidative DNA damage. Vitamin C, the chemical constituent of C. limon has been shown to have high protection abilities against oxidative DNA damage 10. It was therefore envisaged to explore the impact of some plant extracts to assess their DNA damaging/protection potential using pBR322, S. typhi, insect and human DNA.In this study, we have used the plant extracts from A. mexicana,T. peruviana, and C. limonfor the aforesaid purpose.
Materials and Methods
Chemicals
Agarose low EEO (Nuclease and Protease free) (TCI,Tokyo, Japan), Ethidium-bromide, Bromophenol blue (MERCK Pvt. Ltd.,Mumbai, India), Sucrose (Rankem RFCL,New Delhi, India), EDTA, Glacial acetic acid (RFCL, New Delhi, India), Tris base, Ferric chloride (Rankem RFCL, New Delhi, India), Ascorbic acid, Hydrogen peroxide (TCI, Tokyo, Japan)), Sodium Hydroxide, DMSO, ethanol, water, Methanol, Hexane (MERCK Pvt. Ltd.,Mumbai, India) and all other chemicals used were of analytical and molecular grade.
Collection of Plant Materials and Preparation of Extracts
C. limonleaves were collected from home garden, Allahabad. Plant leaves were washed thoroughly under running water, followed by double distilled water (DDW) and then dried in shade on a paper sheet. The dried leaves were crushed as powder. The extract of C. limonleaves was prepared in methanol by Soxhlet method using the leaf powder and methanol in 1:10 ratio (weight/volume). The liquid from the extract was removed by rotatory evaporator using the temperature equivalent to the boiling point (64.7 °C) of methanol to obtain the extract in dry solid state.
A. mexicanaand T. Peruviana leaves were collected from Allahabad, washed thoroughly under running water and dried in shade on paper. Then dried leaves were used for preparation of powder. The extracts were prepared in hexane using Soxhlet method. The ratio of the plant material and the solvent was 1:10 (w/v). The liquid was removed by rotatory evaporator at the temperature equivalent to the boiling point (68°C) of hexane.
Collection and Isolation of DNA
Plasmid pBR322 was purchased from New Biolabs England, insect DNA and Salmonella typhi were obtained in the form of gift from IGBI-New-Delhi. The human DNA was isolated by salting out method from the peripheral blood mononuclear cell separation (PBMNCs) from the human blood. Blood was collected in heparinized vial. The DNA isolation was performed using the procedure described by Miller et al. (1988). The blood sample was added to low salt buffer, mixed thoroughly and centrifuged at 8000xg,250C for 3 min. The supernatant was discarded and again low salt buffer was added to the pellet. This step was repeated till the complete lysis of RBC. To the WBC pellet, high salt buffer, 10% SDS were added and incubated at 370C for 5 min. At the end of incubation, 100µl of 6M NaCl was added and vortexed to precipitate the proteins, followed by centrifugation at 8000xg for 5min. The supernatant was transferred into a new eppendorf tube containing 300 µl of isopropanol, DNA was precipitated, rinsed with 70% ethanol. DNA was dissolved in TAE buffer (pH 8) after air drying.
Preparation of Fenton’s Reagent (FR)
The Fenton’s reagent (FR) was prepared by mixing H2O2, ascorbic acid and FeCl3 in double distilled water (DDW) in the final concentrations of 30mM, 50mM and 80mM, respectively.
Standardization of DNA and Fenton’s Reagent Concentration for DNA Damage Assay
The DNA isolated from the human blood was tested for its purity and used for the DNA damage assay. The concentration of DNA was standardized for this purpose so as to use the suitable amount of DNA in different assays. The 0.8% agarose gel was used in TAE buffer (pH8.0). Ethidium bromide (0.5µg/ml) was added into the chick warm agarose solution. Bromophenol blue (0.25%) as a tracking dye was mixed with the DNA samples. For each experiment, the freshly prepared FR was used. A fixed concentration of DNA (15 ng) was used with increasing concentrations of FR in order to determine its suitable concentration for complete degradation of the DNA. The concentration of FR which completely damaged the DNA was chosen for further experiments.
DNA Damage Assay by Plant Extracts
In order to perform this experiment, different concentrations of plant extract of A. mexicana and T. peruviana were employed to evaluate the extent of DNA damage. The intensity of DNA in the control (untreated) and the experimental sets (treated) was determined by using gel documentation system. The FR was used as a positive control for DNA damage.
DNA Protection Assay by Plant Extracts
Freshly prepared plant extracts were used in each protection assay. In this experiment, the fixed concentrations of FR and damaging plant extract were used followed by addition of varying concentrations of the protecting plant extract (C. limon). The fixed amount of human DNA (15ng) was used in each set of experiments. The intensity of DNA in the control (untreated) and the experimental sets (treated) was determined by using Gel Documentation system. FR was used as a positive control for DNA damage.
Results
Standardization of Plasmid DNA (pBR322) Concentration and DNA Damage by FR
In order to evaluate the efficacy of plant extracts, it was necessary to standardize the concentration of DNA which could be seen clearly under UV rays. It was found to be 15 ng (data not shown). The amount of FR was also standardized which caused complete disappearance of the DNA band. In Figure 1, lanes 1-5 each containing 15ng of DNA was treated with 0, 1, 2, 3, and 4µl of FR, respectively. It displayed DNA damage by FR in concentration dependent manner. The lanes 4 and 5 with 3 and 4 µl of FR displayed complete disappearance of DNA band. Therefore, 3µl of FR was decided to be used for further experiments.
Figure 1.Standardization of plasmid pBR322 concentration and DNA damage by Fenton’s Reagent. Lanes 1-5 contained 15ng of DNA and 0, 1, 2, 3 and 4µl of FR, respectively.
DNA Damage by FR and Protection by C. limon Leaf Extracts
In order to evaluate the effect of C. limon leaves extract on pBR322 against FR induced DNA damage, the plasmid DNA (15 ng) was added with varying concentrations of C. limon leave extract (100, 200, 400, 600, 800, 1000 and 2000 ppm) and 3µl of FR. The results shown in Figure 2 indicated that DNA with FR resulted in its complete damage in term of intensity which, served as a positive control (Figure 2, lane 3). There was gradual increase in the intensity of DNA bands after treatment of DNA with C. limon leaves extract (Figure 2, lanes 4-8). The Figure 2, lane 2 demonstrates the DNA in presence of the solvent (DMSO) in which the plant extract was prepared. There was no effect of the solvent on DNA. Thus, the results proved the ability of plant extract (PE) in preventing DNA from oxidative damage. In order to observe whether higher concentration of this plant extract could be pro-oxidant, we conducted this experiment using its higher concentrations. The results indicated that upon further increasing the PE concentrations from 2000 to 4000, 5000, 6000, 7000, and 9000 ppm, there was increase in the intensity of DNA bands, thereby indicating its DNA protective behavior (data not shown). It did not show any pro-oxidant property at higher concentrations.
Figure 2.DNA protection assay using C. limon leaves extract (diluted in methanol). Lanes 1-3 represent control, lane 1; pBR322 DNA in TAE buffer, lane 2; DNA with solvent (DMSO), and lane 3; contained DNA with FR. Lanes 4-8 contained DNA, FR and different concentrations of PE (in order of DNA, PE and FR). PE concentrations were 100, 200, 400, 600, 800 and 1000 ppm, respectively.
Effect of A. Mexicana on Salmonella Typhi DNA and Protection by C. Limon Leaves Extract
In order to evaluate the effect of A. mexicanaon Salmonella typhi DNA alone and in combination with the C. limon leaves extract, 15ng of DNA was added with varying concentrations of A. mexicana ranging from 90000, 100000, 110000, 120000, 130000, 140000 and 150000ppm and 3µl of FR (able to cause complete DNA damage). The results are shown in Figure 3. The panel A indicated that DNA with FR resulted in its complete damage in term of total loss of intensity which, served as a positive control. (Figure 3, lane 3). There was gradual increase in the intensity of DNA bands after treatment with C. limonleaves extract (Figure 3 lane 4-10). Figure 3, lane 2 demonstrates the DNA in presence of the solvent (DMSO) in which the plant extract was prepared. There was no effect of the solvent on S. typhi DNA, thereby showing the ability of extract in preventing DNA from oxidative damage. The results indicated that there was a gradual decrease in the intensity of DNA bands with increase in the concentration of A. mexicana. The DNA band intensity was reduced to minimum at 150000ppm of A. mexicana extract. In panel B of Figure 3, the effect of C. limon extract on the A. mexicana mediated DNA damage was monitored by adding varying concentrations of the C. limonextract with DNA (15 ng) and A. mexicana (150000ppm). The results shown in Figure 3, panel B indicated that the use of varying concentrations of C. limon extracts (lanes 5-10 corresponding to 2500, 5000, 7500, 10000, 12500 and 15000 ppm, respectively) caused recovery of DNA bands in terms of its increasing intensity. It appears that the extract of C. limon leaves has the potential to protect the DNA from damage caused due to treatment with A.mexicanaleaves extract. Lane 3 of panels A and B of Figure 3 demonstrated complete damage of DNA by FR (3µl) which was used as a positive control. Lane 2 of panel B, Figure 3 demonstrates the DNA in presence of DMSO (the solvent in which the plant extract was prepared).
Figure 3.Effect of A. mexicana on Salmonella typhi DNA and protection by C. limon leaves extract. Panel (A) Lane 1: control i.e. DNA in TAE buffer, lane 2: DNA in DMSO and TAE, lane 3: DNA with FR; lanes 4-10 contained DNA, A. mexicana in increasing concentrations from 90000, 100000, 110000, 120000, 130000, 140000 and 150000ppm, respectively. Panel (B) The lane 1: control i.e. DNA in TAE buffer, lane 2: DNA, DMSO and TAE, lane 3: DNA with FR. Lanes 4-10: DNA, A. mexicana 150000ppm in each and varying concentrations of lemon leaves extract. The order addition was DNA, C. limon leaves extract and A. mexicana extract. The lemon leaves extract concentrations were 2500, 5000, 7500, 10000, 12500 and 15000 ppm, respectively. AM= A. mexicana; CL= C. limon; panel C: representsan estimate of data from panel A, lanes 1, 4 to 10, respectively; panel D: represents an estimate of data from panel B, lanes 3 to 10, respectively.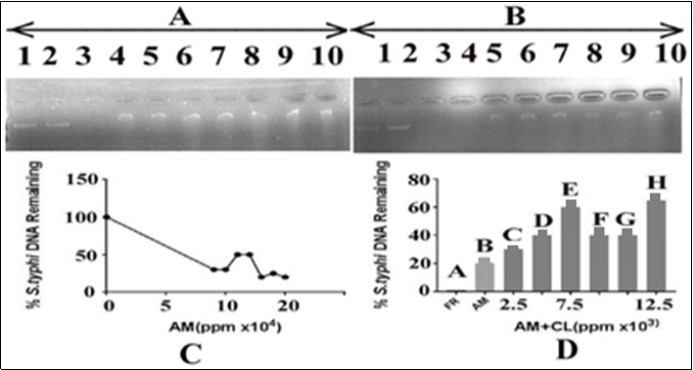
Effect of T. Peruviana on Salmonella Typhi DNA and Protection by C. Limon Leaves Extract
In order to evaluate the effect of T. peruviana leaves extracts on Salmonella typhi DNA alone and in combination with the C. limonleaves extract, 15ng of DNA was added with varying concentrations of T. peruviana leaves extracts i.e. 90000, 100000, 110000, 120000, 130000, 140000 and 150000ppm and 3µl of FR (able to cause complete DNA damage). The results are shown in Figure 4. The panel A indicated that DNA with FR resulted in its complete damage in term of intensity which, served as a positive control (Figure 4, lane 3). The results indicated that there was a gradual decrease in the intensity of DNA bands with increase in the concentration of T. peruviana. The DNA band intensity was reduced to minimum at 150000 ppm T. peruviana leaves extract concentration. In panel B of Figure 4, the effect of C. limonextract on the T. peruviana mediated DNA damage was monitored by adding varying concentrations of the C. limonleaves extract with DNA (15ng) and T. peruviana (150000ppm). The results shown in Figure 4, panel B indicated that the use of varying concentrations (50000, 60000, 70000, 80000, 90000 and 100000 ppm, respectively) of the C. limonleaves extracts caused gradual recovery of DNA bands in terms of its gradual increase in intensity (lanes 5-10). It appears that the extract of C. limonleaves has the potential to protect the DNA from T. peruviana mediated DNA damage; Lane 2 of panel B, Figure 4 demonstrates the DNA in presence of the solvent (DMSO) in which the plant extract was prepared. There was no effect of the solvent on DNA. Lane 3 of the panels A and B of Figure 4 presented complete damage of DNA by FR (3µl) which served as a positive control.
Figure 4.Effect of T. peruviana on Salmonella typhi DNA and protection by C. limon leaf. Panel (A): lane 1control: DNA in TAE buffer; lane 2: DNA, DMSO and TAE buffer; lane 3: DNA with FR;Lanes 4-10 contained DNA, T. peruviana in increasing concentrationsi.e. from 90000, 100000, 110000, 120000, 130000, 140000 and 150000ppm, respectively. (B)The lane 1:control, DNA in TAE buffer, lane 2: DNA, DMSO and TAE buffer; lane 3: DNA with FR. Lanes 4-10 =DNA, T. peruviana150000ppm in each and varying concentrations of lemon leaves extract i.e. 2500, 5000, 7500, 10000, 12500 and 15000 ppm, respectively.TP=T. peruviana; CL=C. limon; panel C: represents an estimate of data from panel A, lanes 1, 4 to 10, respectively; panel D: represents an estimate of data from panel B, lanes 3 to 10, respectively.
Effect of A. Mexicana on Insect DNA and Protection Using Methanolic extract of C. Limon Leaves
In order to evaluate the effect of A. mexicanaextract on insect DNA alone and in combination with the C. limon leaves extract, 15ng of insect DNA was added with varying concentrations of A. mexicana extract ranging from 90000, 100000, 110000, 120000, 130000, 140000 and 150000ppm and 3µl of FR (able to cause complete DNA damage). The results shown in Figure 5, panel A indicated that DNA with FR resulted in its complete damage in term of intensity which, served as a positive control (Figure 5 lane 3). The results indicated that there was gradual decrease in the intensity of DNA bands with increase in the concentrations of extract of leaves of A. mexicana. The insect DNA band intensity was reduced to minimum at 15000 ppm A. mexicana concentration (Panel C). In panel B of Figure 5, the effect of C. lemon extract on the A. mexicanamediated insect DNA damage was monitored by adding varying concentrations of the C. limonleaves extract with insect DNA (15 ng) and a fixed concentration of A. mexicana (15000ppm). The results shown in Figure 5, panel B indicated that the use of varying concentrations of the C. limon extracts (lanes 4-10 corresponding to 2500, 5000, 7500, 10000, 12500 and 15000 ppm, respectively) caused recovery of DNA bands, maximum effect reflected at 75000ppm, in terms of its gradual increase in the intensity. It appears that the extract of C. limon leaves extract has the potential to protect the insect DNA from A mexicana mediated damage (Panel D) ; lane 3 of panels A and B of Figure 5 showed complete damage of insect DNA by FR (3µl) which, served as a positive control. Lane 2 of panel B, Figure 5 demonstrates the insect DNA in presence of the solvent (DMSO) in which the plant extract was prepared. There was no effect of the solvent on insect DNA.
Figure 5.Effect of A.mexicana on insect DNA and protection by using methanolic extract of C. limon leaves. Panel (A), Lane 1: control i.e. DNA in TAE buffer; lane 2: DNA in DMSO; lane 3: DNA with FR; Lanes 4-10 contained DNA, A. mexicana extract in increasing concentrations i.e. 30000, 40000, 50000, 60000, 70000, 80000 and 90000 ppm, respectively. Panel (B), Lane 1:control containing DNA in TAE buffer; lane 2: DNA in DMSO; lane 3: DNA with FR. The lanes 4-10 contained DNA, increasing concentrations of lemon leaves extract i.e. 2500, 5000, 7500, 10000, 12500 and 15000 ppm respectively, and a fixed concentration of A. mexicana extract i.e. 90000. AM=A. mexicana; CL=C. limon; panel C: represents an estimate of data from panel A, lanes 1, 4 to 10, respectively; panel D: represents an estimate of data from panel B, lanes 3 to 10, respectively.
Effect of T. Peruviana on Insect DNA and Protection by C. Limon Leaves Extract
In order to evaluate the effect of T. peruviana on Salmonella typhi DNA alone and in combination with the C. limonleaves extract, 15ng of DNA was added with varying concentrations of T. peruviana i.e. 90000, 100000, 110000, 120000, 130000, 140000 and 150000ppm and 3µl of FR (able to cause complete DNA damage). The results shown in Figure 6, panel A, indicated that DNA with FR resulted in its complete damage as there was total loss of intensity of DNA band, which served as a positive control (Figure 6, lane 3).
Figure 6.Effect of T. peruviana extract on insect DNA and protection by C. limon leaves. Panel (A): lane 1, control containing DNA in TAE buffer; lane 2: DNA in DMSO; lane 3: DNA with FR; lanes 4-10 contained DNA and T. peruviana extract in growing concentrations i.e. 5000,10000,2000,25000,30000,35000, and 40000 ppm, respectively. Panel (B): lane 1, control containing DNA in TAE buffer; lane 2: DNA in DMSO; lane 3: DNA with FR; lanes 5-10 contained DNA, increasing concentrations of lemon leaves extract i.e. 2500, 5000, 7500, 10000, 12500 and 15000 ppm, respectively. and a fixed concentration of T. peruviana (40000ppm). TP=T. peruviana; CL=C. limon; panel C: represents an estimate of data from panel A, lanes 1, 4 to 10, respectively; panel D: represents an estimate of data from panel B, lanes 3 to 10, respectively.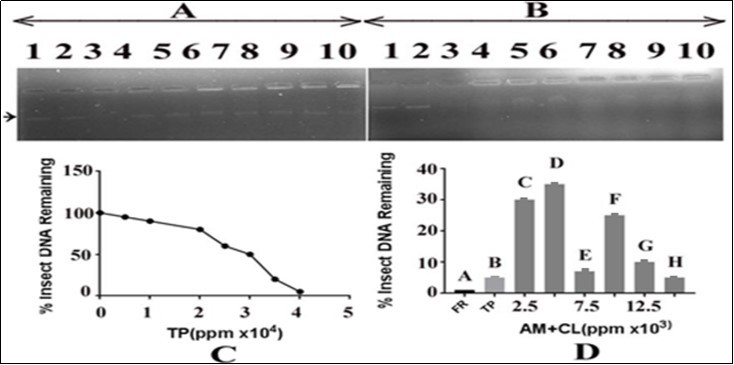
The results indicated that there was a gradual decrease in the intensity of Insect DNA bands with increase in the concentration of T. peruviana. The insect DNA band intensity was reduced to minimum at 150000 ppm of T. peruviana concentration (Panel C). In panel B of Figure 6, the effect of C. limonextract on the T. peruviana mediated DNA damage was monitored by adding varying concentrations of the C. limonextract to a mixture of DNA (15ng) and a fixed concentration of T. peruviana (150000 ppm). The results shown in Figure 6, panel B indicated that the use of varying concentrations of C. limonextracts i.e. 2500, 5000, 7500, 10000, 12500 and 15000 ppm,respectively, caused recovery of insect DNA bands in terms of its growing intensity. However, maximum recovery of the insect DNA was obtained at 5000ppm of C. limon extract, then after it displayed inconsistent effect upon increasing concentrations further. It appears that the extract of C. limonleaves has the potential to protect the DNA from T. peruviana mediated damage (Panel D). Lane 3 of panels A and B of Figure 6 displayed complete damage of DNA by FR (3µl), which served as a positive control. Lane 2 of panel B, Figure 6 demonstrated the DNA in presence of the solvent (DMSO) in which the plant extract was prepared. There was no effect of the solvent on Insect DNA.
Effect of A. Mexicana Leaves Extract on Human DNA and Protection by C. Limon Leaves Extract
In order to evaluate the effect of A. mexicanaon human DNA alone and in combination with the C. limonleaf extract, 15 ng of DNA was added with varying concentrations of A. mexicana ranging from 900000, 100000, 110000, 120000, 130000, 140000 and 150000ppm. The results shown in Figure 7, panel A, Lanes 4-10 indicated that there was gradual decrease in the intensity of DNA bands with increase in the concentration of A. mexicana. The human DNA band intensity was reduced to minimum at 15000 ppm A. mexicana concentration (panel A of Figure 7) showing genotoxic potential of the A. mexicana leaves extract (Panel C). Lane 3 contained DNA (15ng) and 3µl of FR which was able to cause complete human DNA damage and used as a positive control. In panel B of Figure 7, the effect of C. lemon extract on the A. mexicana mediated human DNA damage was monitored by adding varying concentrations of the C. limon extract with DNA (15ng) and a fixed concentration of A. mexicana (150000ppm). The results shown in Figure 7, panel B indicated that the use of varying concentrations of the C. limon extracts (lanes 5-10 corresponding to 2500, 5000, 7500, 10000, 12500 and 15000 ppm, respectively) caused recovery of DNA bands in terms of its increasing intensity. The recovery of DNA, however, did not increase after 7500ppm of the C.limon extract. It appears that the extract of C. limon leaves has the potential to protect the DNA from A mexicana mediated damage (Panel D). Lane 2 of panel B, Figure 7 demonstrated the DNA in presence of the solvent (DMSO) in which the plant extract was prepared. There was no effect of the solvent on human DNA. Lane 3 of panels A and B of Figure 7 displayed complete damage of human DNA by FR (3µl) which, served as a positive control. Lane 4 contained DNA and A mexicana leaves extract (150000ppm).
Figure 7.Effect of A. mexicana on human DNA isolated from blood and protection by methanolic extract of C. limon leaves. Panel (A): lane 1 control containing DNA in TAE buffer; lane 2: DNA in DMSO; lane 3: DNA with FR. Lanes 4-10 contained DNA with hexanolic extract of A. mexicana leaves in increasing concentrations i.e. 5000,10000, 2000, 25000, 30000, 35000 and 40000 ppm, respectively. Panel (B): lane 1; control containing DNA in TAE buffer; lane 2: DNA in DMSO; lane 3: DNA with FR. Lane 4: DNA with A. mexicana hexanolic extract (40000ppm). Lanes 5-10 contain DNA, increasing concentrations of C. limon methanolic leaves extract i.e. 2500, 5000, 7500, 10000, 12500 and 15000 ppm and fixed concentration of A. mexicana hexanolic extract (40000ppm). AM=A. mexicana; CL= C. limon; panel C: represents an estimate of data from panel A, lanes 1, 4 to 10, respectively; panel D: represents an estimate of data from panel B, lanes 3 to 10, respectively.
Effect of T. Peruviana on Human DNA and Protection by C. Limon Leaves Extract
In order to assess the effect of T. peruviana on human DNA alone and in combination with the C. limonleaves extract, 15ng of DNA was added with varying concentrations of T. peruviana i.e. 900000, 100000, 110000, 120000, 130000, 140000 and 150000ppm. The results shown in Figure 8, panel A, Lanes 4-10 indicated that there was gradual decrease in the intensity of DNA bands with increase in the concentration of T. peruviana. The human DNA band intensity was reduced to minimum at 150000 ppm T. peruviana extract concentration (panel A of Figure 8) showing genotoxic potential of the T. peruviana leaves extract. Lane 3 contained DNA (15ng) and 3µl of FR which was able to cause complete human DNA damage and used as a positive control.
In panel B of Figure 8, the effect of C. limonextract on the T. peruviana mediated human DNA damage was monitored by adding varying concentrations of the C. limonextract with DNA (15 ng) and T. peruviana (150000ppm). The results shown in Figure 8, panel B, Lanes 5-10 indicated that the use of varying concentrations of the C. limonextracts (50000, 60000, 70000, 80000, 90000 and 100000 ppm, respectively) caused recovery of DNA bands in terms of its growing intensity. It appears that the extract of C. limonleaves has the potential to protect the human DNA from T. peruviana mediated damage; Lane 2 of panel B, Figure 8 demonstrates the human DNA in presence of the solvent (DMSO) in which the plant extract was prepared. There was no effect of the solvent on human DNA. Lane 3 of panels A and B of Figure 8 displayed complete damage of DNA by FR (3µl) which, served as a positive control. Lane 4 of Panel B contained DNA and T. peruviana (150000ppm).
Figure 8.Effect of T. peruviana on human DNA and protection by C. limon leaves extract. Panel (A) The lane 1; control, DNA with TAE, lane 2; DNA and DMSO, lane 3; DNA with FR and remaining lanes 4-10 contain DNA, T. peruviana in increasing concentrations i.e. 5000, 10000, 2000, 25000, 30000, 35000 and 40000 ppm, respectively. Panel (B) The lane 1; control, DNA with TAE, lane 2; DNA and DMSO, lane 3; DNA with FR, lane 4; DNA, T. peruviana (40000ppm). Remaining lane 5-10; DNA+T. peruviana (40000ppm) and varying concentrations of C. limon leaves extract i.e. 2500, 5000, 7500, 10000, 12500 and 15000 ppm, respectively.
Discussion
Due to the constant exposure of the genomic DNA to various endogenous and environmental agents, it gets damaged and can produce DNA lesions. These lesions can affect the fidelity of DNA replication, and transcription, which can create mutations in important protein coding sequences. The DNA damage can have genotoxic and cytotoxic effects on the cell 1. In order to maintain the integrity of the genome, the prokaryotic and eukaryotic organisms are well equipped with several DNA repair mechanism pathways 11, 12. In general, the lesions in the actively transcribed strand are repaired more rapidly than the lesions in the non-transcribed strand. DNA repair is not only a fundamental cellular process for protecting cells against the damage, but it is also essential to ensure the faithful transmission of genetic information from one generation to the next 1.
Active oxygen species such as superoxide anion radical (O2), hydrogen peroxide (H2O2), and hydroxyl radical (OH). can damage almost all cell components,including DNA, lipid and proteins. Reduction of H2O2 by reducing transition metals results in the formation of hydroxyl and related oxidants via the Fenton reaction. The DNA base damages induced by hypoxanthine / xanthine oxidase or iron and H2O2 have been identified and quantitated. Because iron chelators, EDTA and buffers,including potassium phosphate have been used in these studies, the chemistry of the Fenton reaction has probably been perturbed. Such chelators affect the redox potential of iron and may also scavenge oxygen radicals. Moreover, it has been believed that the Fenton oxidant is reactive as hydroxyl radical, then its diffusion distance from DNA is so short that the iron ion involved in the damage is almost certainly complexes to DNA and not to external ligands. The damage to the bases in DNA and mammalian chromatin by H2O2 and transition metals have been investigated 13. However, in these cases chemical hydrolysis procedures were used that might destroy, alter, or form various products. In a study, the killing of Escherichia coli using H2O2 have been caused due to DNA damage and apparently mediated by iron with NADH as the ultimate reducing agent. Some workers have shown the degradation of the four DNA bases by iron and H2O2 under a variety of in vitro conditions 14. In the present study FR was used as positive control as it displayed very strong DNA damaging potential as has been recorded in this study in a concentration dependent manner.
The mechanisms of oxidative DNA damage have not been elucidated properly. However, the oxidative DNA damage mediated by Fenton reactions has been reported to be the most acceptable hypothesis. Free radicals, commonly known as reactive oxygen species (ROS), contain one or more unpaired electrons in their outermost orbital. Excessive production of free radicals results in depletion of antioxidants in vivo and causes an imbalance between free radicals and the antioxidant defenses of the body, which results into the generation of oxidative stress mediated DNA damage. The 8-hydroxydeoxyguanosine (8-OHdG) is the most common biomarker of oxidative DNA damage by chemical carcinogens in which oxidation of a specific base i.e. guanosine in DNA causesan increase in the level of hydroxydeoxyguanosine (8-OHdG). These oxidative chemical species may cause deamination of cytosine converting it into uracil or may remove an individual base generating apurinic / apyrimidinic (AP) sites in DNA 15, 16, 17.
On the other hand, there are some plant products which have shown to possess DNA damaging potential, most of them are used in the treatment of cancer. Recent findings suggested an active role of nicotine, a major tobacco alkaloid present which induces carcinogenesis. Nicotine exhibits tumor promoting potential by causing DNA damage in different human epithelial and non-epithelial cells 18. Another alkaloid, sanguinarine, isolated from a wild plant, A. mexicana, has been shown to cause chromosomal aberration, micronucleus formation and DNA damage by comet assay in mouse models in vivo system. Sanguinarine is reported to inhibit the activity of epidermal histidase leading to the increase in the levels of keratin formation and tumor promotion. The DNA damaging, cytotoxic, anti-cancer potential of A. mexicanaand T. peruviana have been illucidated by some workers 19, 20, 21. In the present investigation, the DNA damaging properties of A. mexicana and T. peruviana leaves extracts were observed against pBR322 plasmid, S. typhi, insect and human DNA. The results from the present study have indicated that the A. mexicana and T. peruviana leaves extracts were able to cause significant DNA damage in vitro in a concentration dependent manner.
As nature provided us toxic plant products, it has also given antioxidants to us, which are usually the free radicals neutralizing and reducing agents such as vitamins, carotenoids, flavones, flavonoids and polyphenols, which scavenge the reactive oxygen species (ROS) and inhibit the chain reaction initiated by them 22. DNA damage inhibition by the methanolic extract of C. carandas leaves have been demonstrated 7. The aqueous extract of Ganoderma lucidum occurring in southern part of India has demonstrated significant antioxidant property and revealed the potential to protect DNA from radiation mediated damage. These findings were suggestive of the possibility of using the medicinal extracts containing flavones, polyphenols, flavonoids, terpenes, tannins and alkaloids as alternative therapeutics in the treatment of cancer 23. Arecoline, an alkaloid constituent of Areca nut has been used in thetreatment of oral and pharyngeal cancers 24. In addition to their free radical quenching potential, the plant products help chelate heavy metals and protect the DNA from damage. Also, some vitamins such as Vitamin C and E have been shown as quenchers of free radicals and therefore, they inhibit the DNA damaging properties of xenobiotics in the living cells 15. The results of the present in vitro study indicated that the extracts of leaves of T. peruviana and A. mexicana were causing damage to DNA isolated from different living systems whereas C. limon was able to protect the DNA from damage in the concentration dependent manner.
Conclusion
The phytochemicals present in different plant species are known to exhibit varied medicinal properties depending on their source, structure and function. It was interesting to observe the results from the present study carried out in vitro to assess the impact of extracts of leaves from four different plants A. mexicana, T. peruviana, and C. limon. It was observed that T. peruviana and A. mexicana leaves extracts were causing damage to DNA isolated from different living systems. However, the extract from C. limonleaves was able to protect DNA from damage by both the FR and the extracts from A. mexicana, T. peruviana, in the concentration dependent manner. At this stage, we are not able to comment on how exactly it is happening. However, further research is needed to isolate and extensively characterize the specific plant molecules responsible for their DNA damaging/protecting potentials so as to delineate their associated mechanisms of specific actions.
Acknowledgments
NS, VKG,PKD,NS and AK are grateful to the University Grant Commission, New Delhi, for providing financial assistance in the form of a Research Fellowship. Authors acknowledge UGC-SAP and DST-FIST for support to Department of Biochemistry, University of Allahabad, Allahabad, India. VKG acknowledges ICMR- New Delhi for financial assistance in the form of senior research fellowship.
References
- 1.Tuteja N, Singh M B, Misra M K, Bhalla P L, Tuteja R. (2001) Molecular mechanisms of DNA damage and repair: Progress in plants. , Crit Rev Biochem Mol 36(4), 337-397.
- 2.Bohr V A, Stevnsner T, Souza-Pinto N C. (2002) Mitochondrial DNA repair of oxidative damage in mammalian cells.Gene. 286(1), 127-134.
- 3.Floyd R A, Watson J J, Wong P K, Altmiller D H, Rickard R C. (1986) Hydroxyl free radical adduct of deoxyguanosine: sensitive detection and mechanisms of formation. Free Radic Res Commun.1(3):. 163-172.
- 4.Shimoda R, Nagashima M, Sakamoto M, Yamaguchi N, Hirohashi S. (1994) Increased formation of oxidative DNA damage,8- hydroxydeoxyguanosine, in human livers with chronic hepatitis. , Cancer Res 54(12), 3171-3172.
- 5.Kolodner R D, Putnam C D, Myung K. (2002) Maintenance of genome stability in Saccharomyces cerevisiae. , Science 297(5581), 552-557.
- 6.Azqueta A, Collins A. (2016) Polyphenols and DNA Damage: A Mixed Blessing. , Nutrients. Dec 8(12), 785.
- 7.Verma K, Shrivastava D, Kumar G. (2015) Antioxidant activity and DNA damage inhibitionin vitroby a methanolic extract ofCarissa carandas(Apocynaceae) leaves. , J Taibah Univ Sci 9(1), 34-40.
- 8.Brahmachari G, Gorai D, Roy R. (2013) chemical and pharmacological aspects. RevistaBrasileira de Farmacognosia. 23(3), 559-575.
- 9.Ramos-Silva A, Tavares-Carreón F, Figueroa M, S De la Torre-Zavala, Gastelum-Arellanez A. (2017) Anticancer potential of Thevetia peruviana fruit methanolic extract. , BMC Compl Alternative Med 17(1), 241.
- 10.Makni M, Jemai R, Kriaa W, Chtourou Y, Fetoui H. (2018) . Citrus limon from Tunisia: Phytochemical and Physicochemical Properties and Biological Activities, BioMed Res Int 1-10.
- 11.Hoeijmakers J H. (2001) Genome maintenance mechanisms for preventing cancer. , Nature 411(6835), 366-74.
- 12.Sancar A, Lindsey-Boltz L A, Unsal-Kacmaz K, Linn S. (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. , Annu Rev Biochem 73, 39-85.
- 13.Lee D H, O'Connor T R, Pfeifer G P. (2002) Oxidative DNA damage induced by copper and hydrogen peroxide promotes CG-->TT tandem mutations at methylated CpG dinucleotides in nucleotide excision repair-deficient cells. , Nucleic Acids Res 30(16), 3566-3573.
- 14.Luo Y, Henle E S, Linn S. (1996) Oxidative Damage to DNA Constituents by Iron-mediated Fenton Reactions. , J Biol Chem 271(35), 21167-21176.
- 15.Singh N, Kumar A, Gupta V K, Sharma B. (2018) . Biochemical and Molecular Bases of Lead-Induced Toxicity in Mammalian Systems and Possible Mitigations,Chem Res Toxicol.31 10, 1009-1021.
- 16.Singh N, Sharma B. (2018) Biotoxins Mediated DNA Damage and Role of Phytochemicals in DNA Potection. , Biochem Mol Biol 4(1), 1-5.
- 17.Singh N, Sharma B. (2019) Role of Toxicants. in Oxidative Stress Mediated DNA Damage and Protection by Phytochemicals”. EC Pharmacol Toxicol 7(5), 325-330.
- 18.Ginzkey C, Kampfinger K, Friehs G, Köhler C, Hagen R et al. (2009) Nicotine induces DNA damage in human salivary glands. , Toxicol Letters 184(1), 1-4.
- 19.Ansari K M, Dhawan A, Khanna S K, Das M. (2005) In vivo DNA damaging potential of sanguinarine alkaloid isolated from argemone oil using alkaline comet assay. Food Chem Toxicol. 43(1), 147-153.
- 20.Das M, Ansari K M, Dhawan A, Shukla Y, Khanna S K. (2005) Correlation of DNA damage in Epidemic Dropsy patients to carcinogenic potential of argemone oil and isolated sanguinarine alkaloid in mice. , Int J Cancer 117(5), 709-717.
- 21.Ansari K M, LKS Chauhan, Dhawan A, Khanna S K, Das M. (2004) Unequivocal evidence of genotoxic potential of argemone oil in mice. , Int J Cancer 112(5), 890-895.
- 22.Vauzour D, Rodriguez-Mateos A, Corona G, Oruna-Concha M J, JPE Spencer. (2010) Polyphenols and human health: prevention of disease and mechanisms of action. , Nutrients 2(11), 1106-1131.
Cited by (3)
- 1.Munir Neelma, Khilji Sheza Ayaz, Rasool Sajal, Khalil Anam, Sajid Zahoor Ahmad, et al, 2025, Phytochemical Analysis, Antibacterial, and Antitumor Potential of Hibiscus rosa-sinensis Linn, Scientifica, 2025(1), 10.1155/sci5/2722306
- 2.Arowoogun Jeremiah, Akanni Olubukola O., Adefisan Adedoyin O., Owumi Solomon E., Tijani Abiola S., et al, 2021, Rutin ameliorates copper sulfate‐induced brain damage via antioxidative and anti‐inflammatory activities in rats, Journal of Biochemical and Molecular Toxicology, 35(1), 10.1002/jbt.22623
- 3.Behl Tapan, Kaur Gagandeep, Sehgal Aayush, Zengin Gokhan, Singh Sukhbir, et al, 2022, Flavonoids, the Family of Plant-Derived Antioxidants Making Inroads into Novel Therapeutic Design Against Ionizing Radiation-Induced Oxidative Stress in Parkinson’s Disease, Current Neuropharmacology, 20(2), 324, 10.2174/1570159X19666210524152817
